SNP Molecular Markers Related to Cotton Flower Spotting Traits and Its Application
A technology related to molecular markers and molecular markers, which is applied in the field of plant molecular biology and achieves the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] 1. Experimental materials and group construction
[0019] Cotton with flower basal spot material is crossed with conventional non-flower basal spot cultivars to obtain F1, and F1 is self-crossed to obtain F2, which is used as a genetic population for research. In order to control the systematic error of the field experiment, all materials were planted in the same field to ensure consistent environmental conditions.
[0020] 2. Investigation on the basal spots of cotton flowers
[0021] The investigation of the basal spot traits of each individual plant in the F2 population is in line with Mendel's law of segregation. Controlled by dominant genes.
[0022] 3. BSR high-throughput sequencing
[0023] Based on the above genetic analysis results, samples were taken from individual plants with and without basal spots in the F2 population, and a mixed pool of progenies with and without basal spots was constructed for BSR high-throughput sequencing. A total of 21854 polymor...
Embodiment 2
[0039] The present embodiment provides a method for detecting cotton flower basal spots using cotton flower basal spot traits related SNP molecular markers, comprising the following steps:
[0040] (1) Utilize the conventional plant tissue DNA extraction method to extract the cotton tissue DNA to be detected;
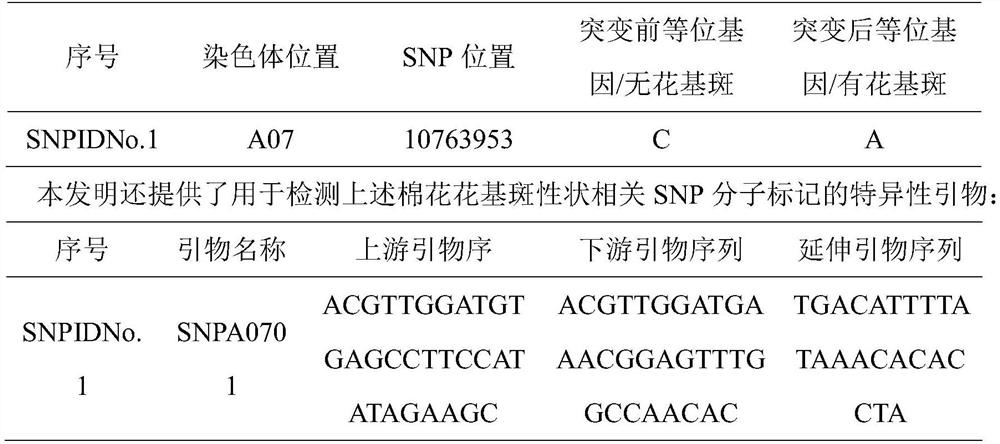
[0041](2) Using MASSARRAY technology, design specific primers according to the SNP molecular marker, the sequence is as follows:
[0042] Upstream primer: ACGTTGGATGTGAGCCTTCCATATAGAAGC
[0043] Downstream primer: ACGTTGGATGAAACGGAGTTTGGCCAACAC
[0044] Extension primer: TGACATTTTATAAACACACCTA
[0045] (3) Use the cotton tissue DNA as a template, and use the designed specific primers to perform PCR amplification
[0046] Take 1 μL of DNA solution with a concentration of 10ng / μL for PCR amplification. The PCR reaction solution should contain Water, HPLC grade 927.5 μL, 10x PCR Buffer with 15mM MgCl 2 331.25 μL, 25 mM MgCl 2 172.25μL, 53μL of 25mM dNTP Mix, 530μL o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com