Cpf1 based transcription regulation systems in plants
A plant and transcription factor technology, applied in the direction of plant regeneration, plant genetic improvement, botany equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
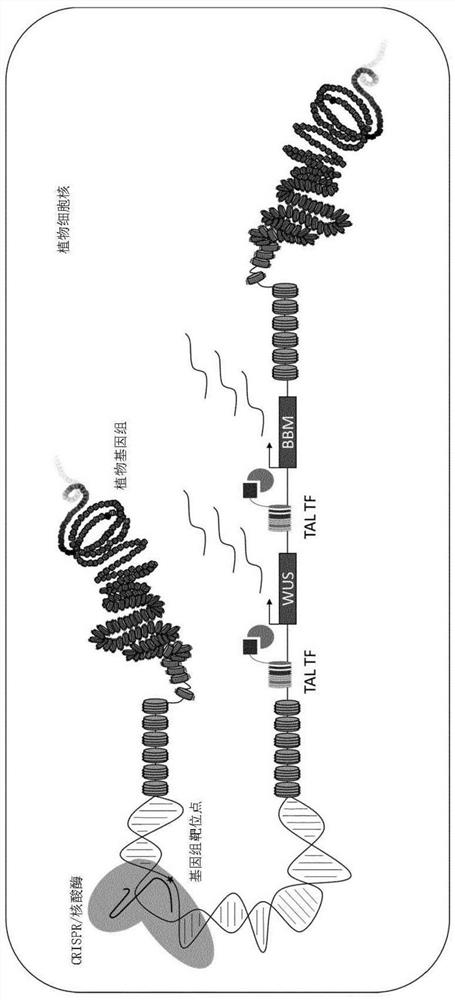
[0383] Example 1: TAL Transcription Factors for Transient Expression of Endogenous Morphogenetic Genes in Maize (Zm)
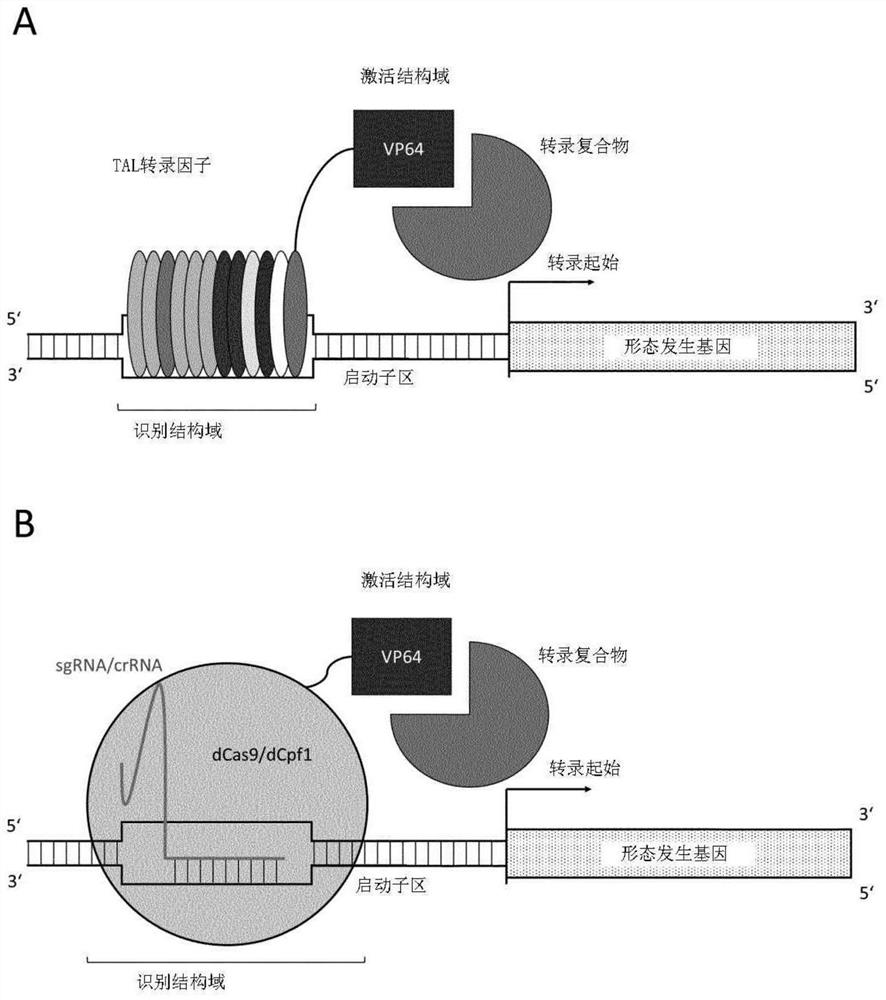
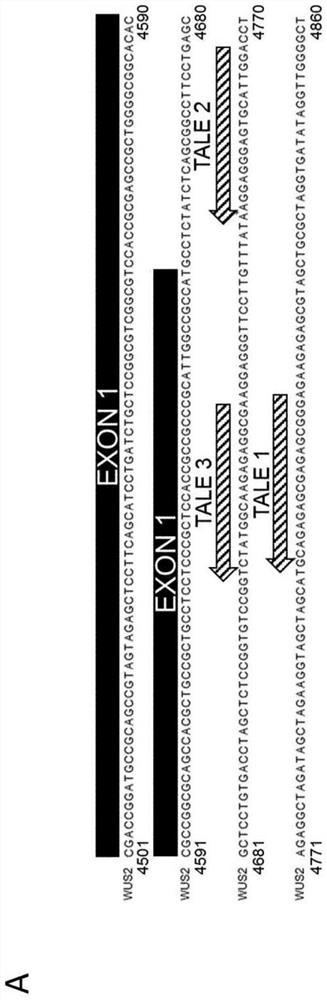
[0384] In one embodiment, commercially designed and constructed TAL transcription factors are used to temporarily enhance the expression of BBM and WUS. The TAL transcription factor is designed to combine with the regulatory region of about 24 bp shown in SEQ ID NO:95, 109-147, 270-272 of BBM and / or the regulatory region of about 18 bp shown in SEQ ID NO:96, 148-190 of WUS (See Figures 3A and 3B). The TAL transcription factor recognition domain of BBM comprises the sequence shown in SEQ ID NO: 13-51, and / or the TAL transcription factor recognition domain of WUS comprises the sequence shown in SEQ ID NO: 52-94.
[0385] TAL effector sequences can be designed and cloned, and the activation domain of herpes simplex virus (VP16 or tetrameric VP64) can be added to the construct in a fusion protein-like manner.
[0386]Transient induction of expression was first t...
Embodiment 2
[0389] Example 2: Fusion protein between non-functional CRISPR-nuclease and activation domain for transient expression of endogenous morphogenetic genes in maize
[0390] Similar to Example 1, constructs were designed for transient delivery, in this case expressing dCas9 (PAM variant available) or dCpf1 (PAM variant available) as a protein with an activation domain (e.g., VP16 or VP64). fusion protein. Potential target sites / regulatory regions include: the Cas9 target sequence shown in SEQ ID Nos: 97-99 of ZmBBM; the Cpf1 target sequence shown in SEQ ID Nos: 100-102 of ZmBBM; the SEQ ID NOs of ZmWUS2: 103- Cas9 target sequence shown in 105; Cpf1 target sequence shown in SEQ ID Nos: 106-108 of ZmWUS2.
[0391] Based on the regulatory regions of CRISPR / dCas9 and CRISPR / dCpf1 described above, a CRISPR-based transcription factor system having a recognition domain comprising the sequence shown in SEQ ID NO: 1-12 can be designed and commercially obtained.
[0392] Transient induct...
Embodiment 3
[0394] Example 3: Replacement of Activation Domains to Optimize Expression of Morphogenetic Genes
[0395] This example aims to test the behavior of different aforementioned activation domains in a systematic manner. This was sufficient to assess their effect on the expression levels of ZmWUS and ZmBBM. As noted above, different STFs for a specific target gene of interest may contain different activation and recognition domains and other elements. Therefore, it is very suitable for designing different STFs for one target and the same target, so as to finally define the optimal STF for regulating the gene of interest.
[0396] The native activation domains of the TAL effector genes of Xanthobacterium oryzae are the most obvious activation domains to use with TAL transcription factors, and also represent an activation domain that can be used alone or in combination according to various aspects of the invention but has also been used in Other set activation domains. They belon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com