Identification method of Tan sheep-derived components
A source ingredient, Tan sheep technology, applied in the fields of molecular biology and food testing, to achieve the effect of enhancing the overall brand value, promoting the healthy and rapid development of the industry, and reducing the number of non-coding areas
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 SNP site combination ability to distinguish Tan sheep breeds
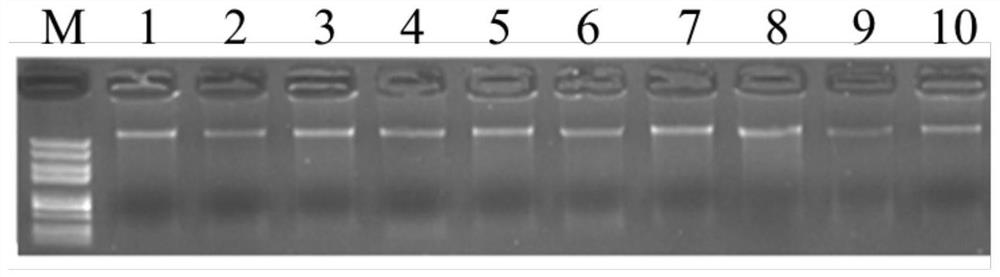
[0056] The genomic DNA of 100 Tan sheep and 44 non-Tan sheep was extracted by CTAB method, and the quality was checked by agarose gel electrophoresis. figure 1 ). The DNA that passed the quality inspection was amplified and sequenced by PCR, and a total of 144 mitochondrial gene sequences of sheep were obtained. At the same time, 19 non-Tan sheep mitochondrial genes were downloaded from the NCBI website (https: / / www.ncbi.nlm.nih.gov / ), and a total of 163 mitochondrial DNA sequences of Tan sheep and non-Tan sheep samples were obtained.
[0057] The collected 163 mitochondrial gene information was processed by Python program and Mega software to construct the Tan sheep identification method. The loci of SNP loci that are conserved in Tan sheep species and with high polymorphism in other breeds were selected, and 35 loci were selected from 399 loci as characteristic markers for identifying Tan sheep...
Embodiment 2 3
[0060] Example 2 Establishment and specificity of triple and quadruple RT-PCR reaction systems
[0061] 1. SNP site primer design
[0062] Use Oligo 7.0 software to design PCR amplification primers for the SNP site to be tested (Table 1), and hand them over to Yingweijieji Company for synthesis.
[0063] 2. Establishment of RT-PCR reaction system
[0064] Combine primers according to reaction system A: 12.5 μL of Premix master mix, 0.3-1.5 μL of 5 μM Primer 1F and Primer 1R each, 0.3-1.5 μL of 5 μM Primer 2F and Primer 2R each, 0.3-1.5 μL of 5 μM Primer 3F and Primer 3R each, 5μM Primer 4F and Primer 4R each 0.3-1.5μL, 0.5ng / μL template DNA 1μL, deionized water to make up to 25μL;
[0065] Reaction system B: 12.5μL of Premix master mix, 0.3-1.5μL each of 5μM Primer 5F and Primer 5R, 5μM Primer 6F and Primer 6R each 0.3-1.5μL, 5μM Primer 7F and Primer 7R each 0.3-1.5μL, 0.5ng / μL template DNA 1 μL, deionized water to make up to 25 μL.
[0066] Under the condition of ensuring s...
Embodiment 3
[0074] Example 3 Sample Detection and Method Accuracy Investigation
[0075] 1. Collect samples and prepare templates
[0076] 60 DNA samples from Tan sheep and 60 non-Tan sheep samples were extracted by CTAB method. According to the DNA extraction method in Reference Example 1, the DNA of Tan sheep and non-Tan sheep were extracted respectively, and the DNA concentration and purity were calculated. The concentration of DNA samples was diluted to 0.5ng / μL and used as detection template.
[0077] 2. Validation of triple and quadruple RT-PCR reactions
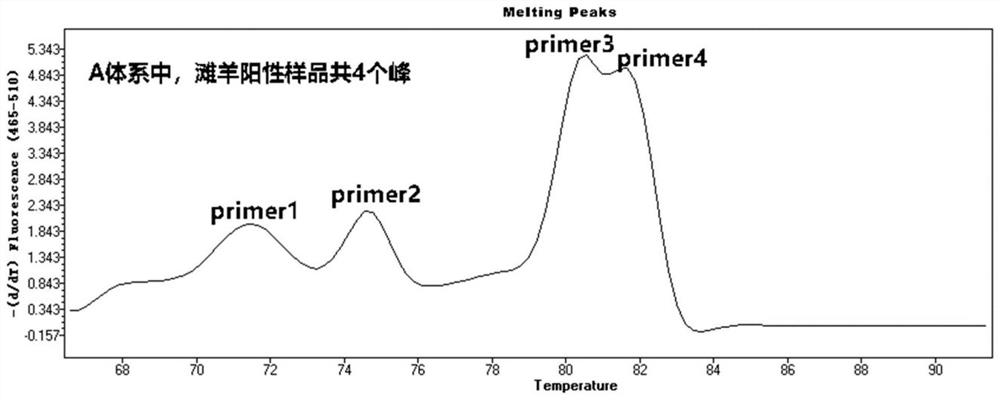
[0078] Referring to the RT-PCR reaction process in Example 2, 120 samples of Tan sheep and non-Tan sheep DNA templates were simultaneously or separately subjected to triple and quadruple RT-PCR melting curve analysis in two reaction wells, and Tm values were calculated and counted. The number of melting curve peaks.
[0079] 3. Sample verification
[0080] Determination criteria for Tan sheep: 60 Tan sheep and 60 non-Tan sh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com