Application of a Fluorescent Probe with Dual Fluorescent Emissions in Detecting RNA
A fluorescent probe and fluorescent emission technology, applied in the application field of fluorescent probes and RNA detection, can solve the problems of low detection efficiency, low accuracy, low RNA detection specificity, etc., and achieve good linear correlation, The effect of specific detection and wide detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1: The ability of fluorescent probe TPBT to distinguish single-stranded RNA and double-stranded RNA
[0047] figure 2 Ribonucleotide A in different concentrations of 0, 0.2, 0.4, 0.6, 0.8, 1.0 μM is added to SYBR Green I aqueous solution 50 with U 50 The composition of the double helix structure (A+U) 50 The fluorescence titration spectrum. image 3 Add 0, 0.2, 0.4, 0.6, 0.8μM different concentrations of single-stranded ribonucleotide A to the aqueous solution of SYBR Green I 50 Fluorescence titration spectra. Figure 4 It is in 10μM TPBT and excess polyanionic heparin sodium (200-2000μg·mL -1 ) to the aqueous solution composed of 0, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, 2.0, 2.2, 2.4, 2.6, 2.8, 3.0μM ribonucleotide A in different concentrations 50 with U 50 The composition of the double helix structure (A+U) 50 Fluorescence titration spectra. Figure 5 10μM TPBT with excess polyanion sodium heparin (200-2000μg·mL -1 ) to the aqueous solutio...
Embodiment 2
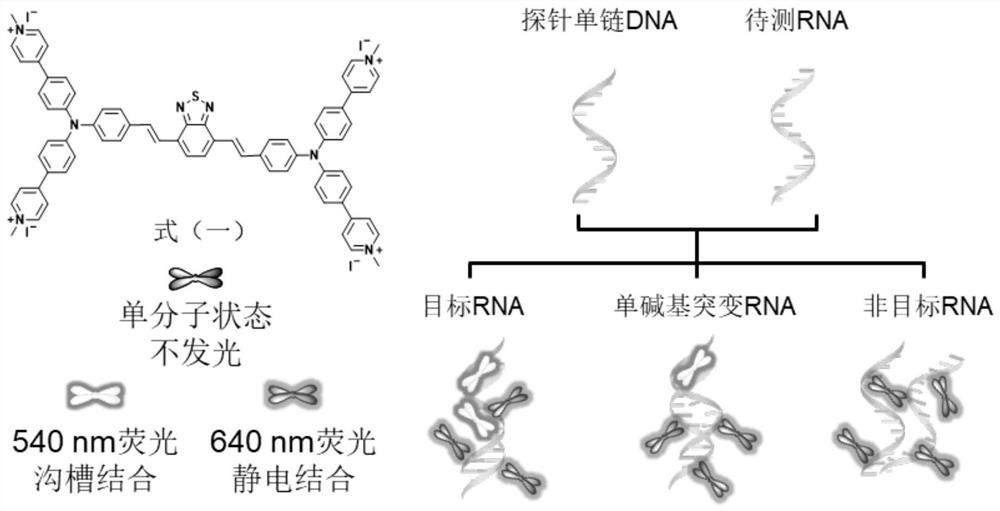
[0050] Embodiment 2: TPBT detects HIV RNA fragment and single base mutation thereof
[0051] 200 μg·mL was added dropwise to an aqueous solution of TPBT at a concentration of 10 μM -1 Heparin sodium, so that its 640 nm fluorescence emission intensity reaches balance, and the detection reagent of TPBT and heparin sodium is formed for use. SEQ ID NO 1, SEQ ID NO 2, SEQ ID NO 3 and SEQ ID NO 4 are HIV RNA, HIV RNA with single nucleotide mutation (mis HIV) and non-target RNA (nonHIV) and probe single-stranded DNA base sequence. Preparation of RNA-DNA double helix structure: Dissolve single-stranded probe DNA and RNA to be tested in ultrapure water to prepare a 0.5mM mother solution, mix probe DNA with target HIV RNA, single-base mutant mis HIV RNA and non- The target non-HIV RNA was mixed at 37°C and 225rad / min for 5-10min to obtain the corresponding RNA-DNA pair. Then the corresponding RNA-DNA pairs were titrated into the detection reagents of TPBT and sodium heparin respectiv...
Embodiment 3
[0053] Embodiment 3: Commercial dye SYBR Green I detects HIV RNA fragment and single base mutation thereof
[0054] Figure 10-13 In embodiment 3, commercial dye SYBR Green I detects the experiment of target RNA and single base mutation thereof. The DMSO mother solution of SYBR Green I at 10000× was diluted 10000 times with ultrapure water for use as a detection reagent. The preparation of the RNA-DNA double helix structure is the same as in the above-mentioned Example 3, and then the corresponding RNA-DNA pairs are respectively titrated into the detection reagent of SYBR GreenI, and the fluorescence intensity thereof is detected. Figure 10 It is the fluorescence spectrum of the RNA-DNA paired with HIV RNA and probe DNA in the detection reagent of SYBR Green I. Figure 11 It is the fluorescence spectrum of the RNA-DNA paired with the probe DNA titrated mis HIV RNA in the detection reagent of SYBR Green I. Figure 12 It is the fluorescence spectrum of the RNA-DNA paired wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com