An artificial noncoding RNA module that enhances microbial nitrogen fixation
An artificial nitrogen fixation technology, applied in the biological field, can solve the problems of low nitrogen fixation efficiency and limited application, and achieve the effect of enhancing nitrogen fixation ability and nitrogenase activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Construction of AneR fusion expression vector
[0044] (1) Experimental method:
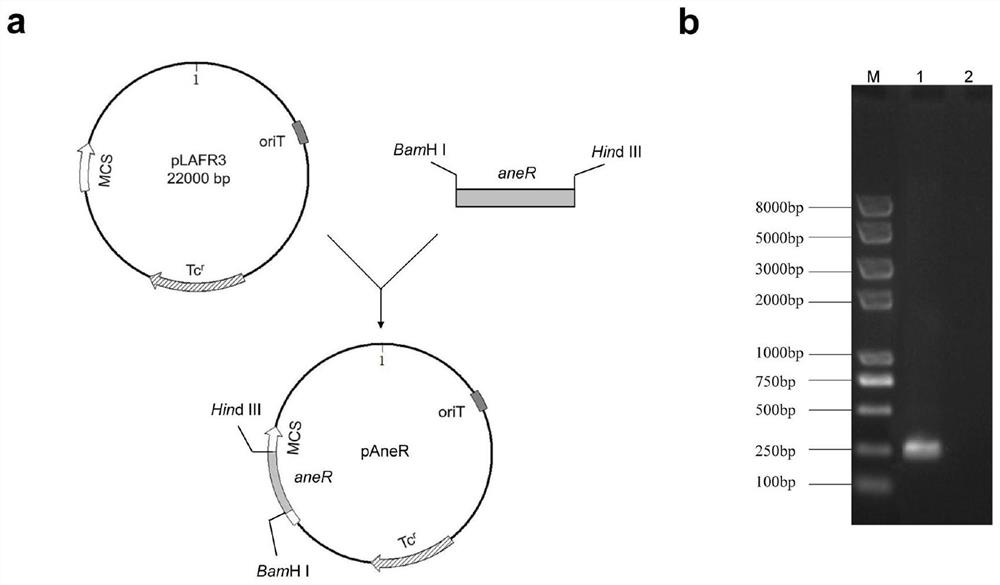
[0045]Firstly, an artificial RNA module AneR with a full length of 505 bp was synthesized by artificial chemical synthesis, and its expression was controlled by an artificial promoter element with a size of 360 bp. Then the artificial RNA module AneR and the expression vector pFLAα3 were subjected to Bam HI and Hind III double enzymes respectively. Then, T4 DNA ligase was used to insert the recovered AneR fragment into the multi-cloning site of pFLAα3. Finally, the AneR fusion expression vector pAneR was obtained by PCR sequencing verification. The expression vectors were transferred into three different nitrogen-fixing microbial chassis (P. stutzeri, K. pneumoniae, and A. vinelandii) by tri-philic binding or electrical stimulation. , and obtained three recombinant nitrogen-fixing engineering strains.
[0046] (2) Experimental results:
[0047] The full-length nucleic acid seq...
Embodiment 2
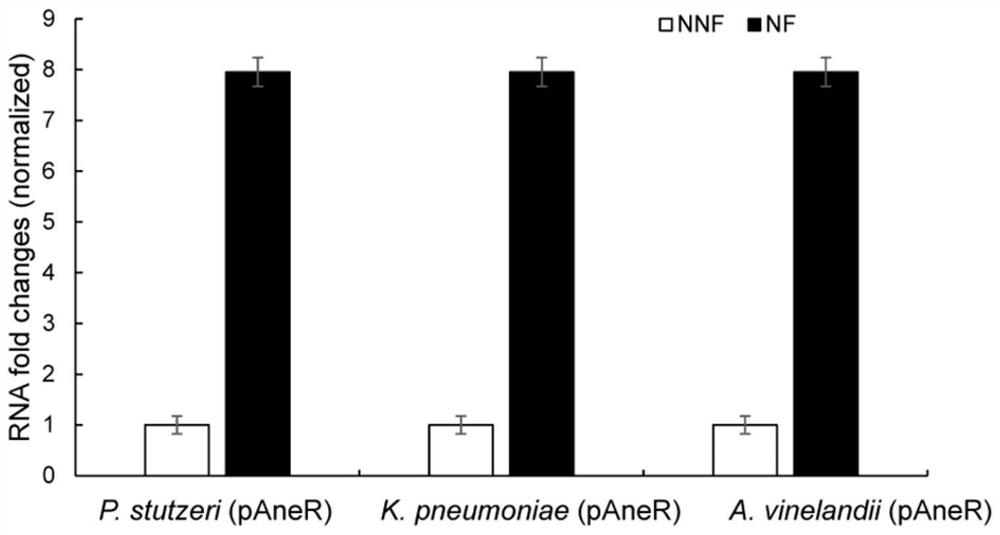
[0050] Example 2 Expression analysis of artificial RNA module AneR in recombinant engineering strains under nitrogen fixation conditions
[0051] (1) Experimental method
[0052] 1. The recombinant strains P.stutzeri (pAneR), K.pneumoniae (pAneR) and A.vinelandii (pAneR) were activated in LB liquid medium and cultured at 30°C overnight;
[0053] 2. Centrifuge the cells at 4000rpm / 10min the next day, and wash the cells twice with normal saline;
[0054] 3. Suspend the bacteria with normal saline and adjust the OD 600 ≈1.0;
[0055] 4. The bacteria were cultured under normal conditions and nitrogen-fixing conditions, respectively, and the OD was adjusted. 600 ≈0.5;
[0056] 5. After the culture medium was shaken and cultured at 30°C for 0.5h, the cells were collected by centrifugation at 8000rpm for 5min;
[0057] 6. Use Promega Mass Extraction Kit Z3741 to extract total bacterial RNA, and reverse the single-stranded DNA (cDNA) with the same amount of sample RNA;
[0058] ...
Embodiment 3
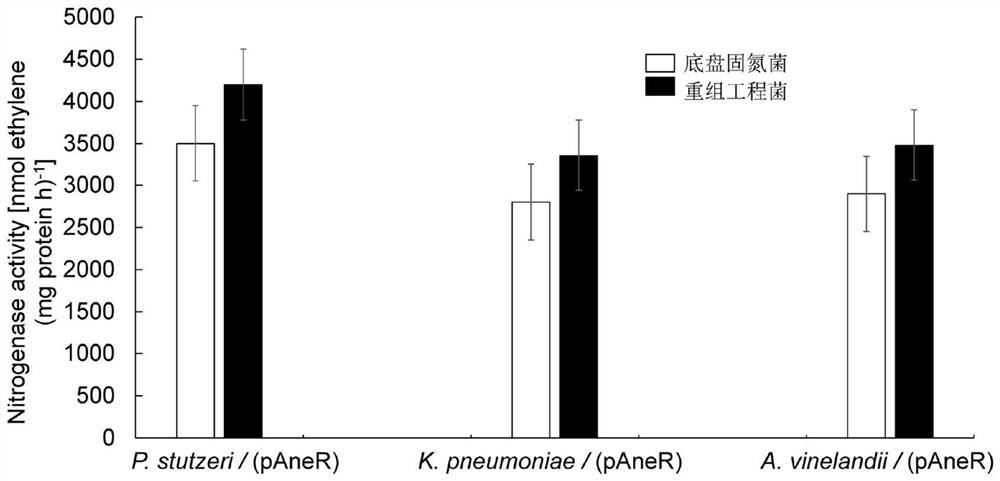
[0063] Embodiment 3 The nitrogenase activity assay of recombinant nitrogen-fixing engineering strain
[0064] (1) Experimental method
[0065] The nitrogenase activity of the recombinant engineering strain was determined by the internationally recognized acetylene reduction method. The specific steps are as follows:
[0066] 1. The chassis strains P.stutzeri, K.pneumoniae and A.vinelandii and the recombinant strains P.stutzeri (pAneR), K.pneumoniae (pAneR) and A.vinelandii (pAneR) were activated in LB liquid medium at 30°C overnight nourish;
[0067] 2. Centrifuge the cells at 4000rpm / 10min the next day, and wash the cells twice with normal saline;
[0068] 3. Suspend the bacteria with normal saline and adjust the OD 600 ≈1.0, the cells were cultured under the following conditions:
[0069] P. stutzeri and recombinant strain P. stutzeri (pAneR)
[0070] (1) Transfer the bacterial suspension to a grinding-mouthed conical flask equipped with K medium (without N), and adjust...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com