Method for obtaining substrate sequence efficiently bound in ADAR protein cells, substrate sequence and application
A protein and substrate technology, which is applied in the field of acquisition, fixed-point targeted RNA editing, and positioning of substrate sequence information for efficient binding of ADAR proteins in cells. The clinical application and the low proportion of chimeras can achieve the effects of shortening the experimental time, increasing the yield, and increasing the number of constructions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Construction of ADAR1 Protein High Affinity Substrate Sequence and RNA Level Targeted Editing in HEK293 Cell Line
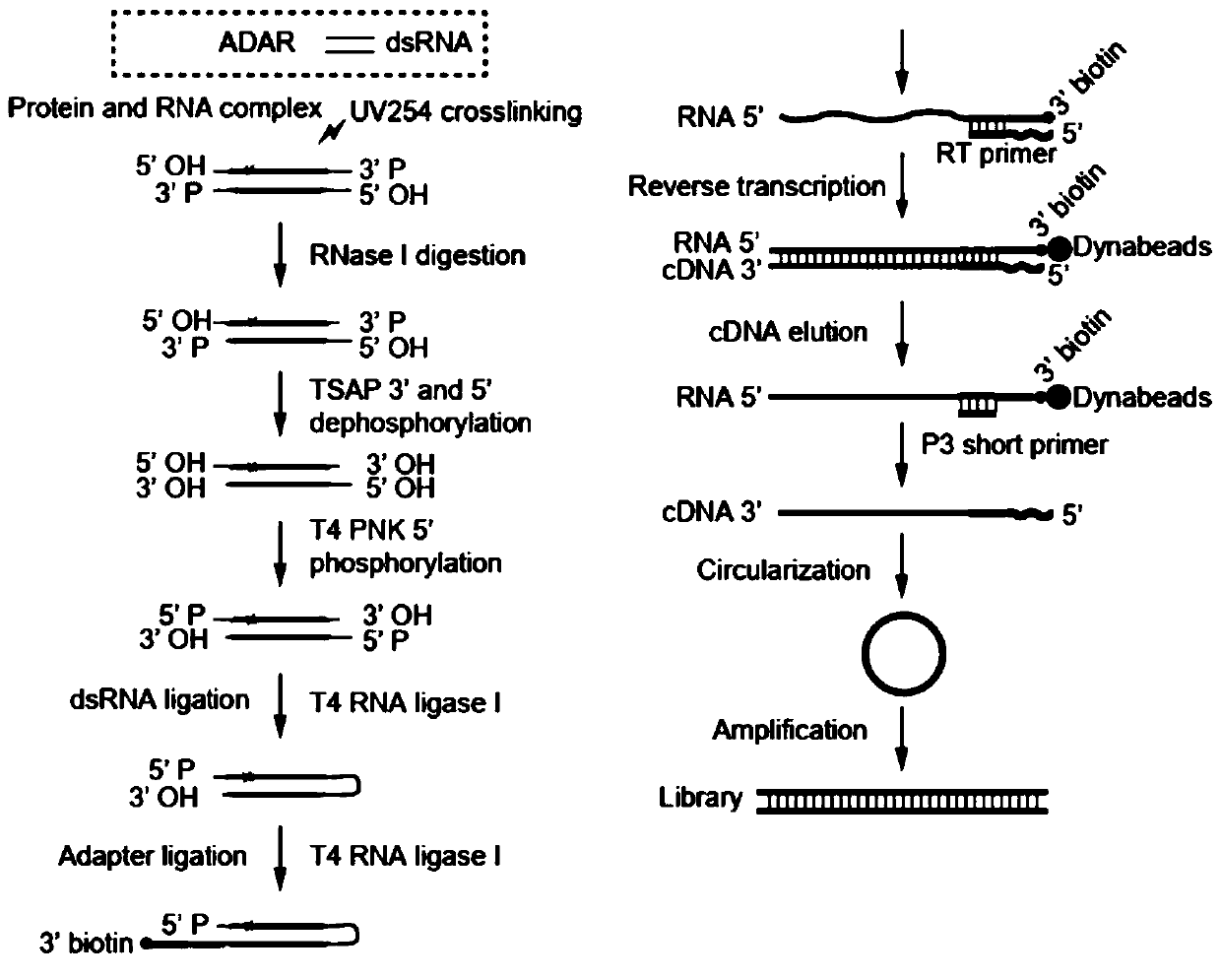
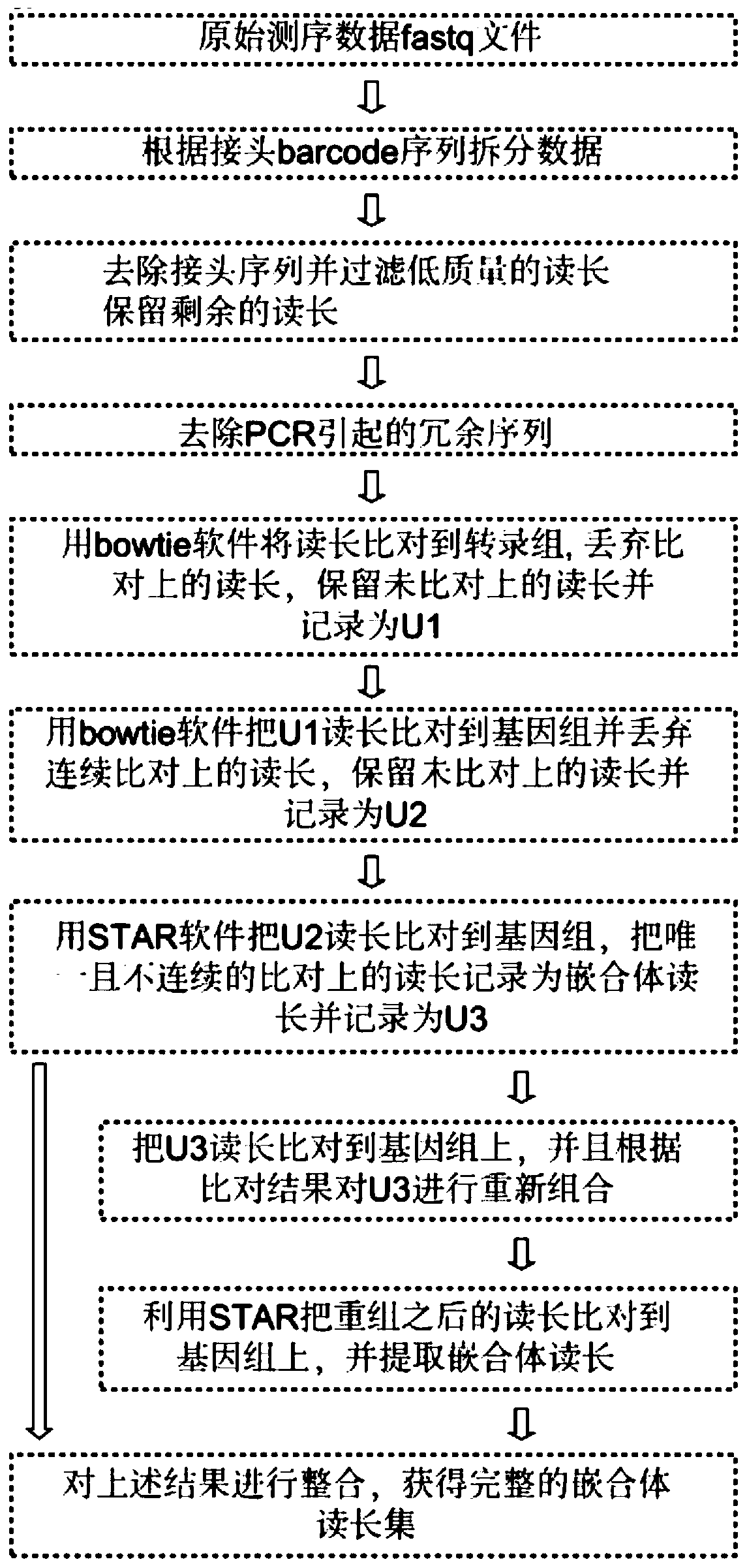
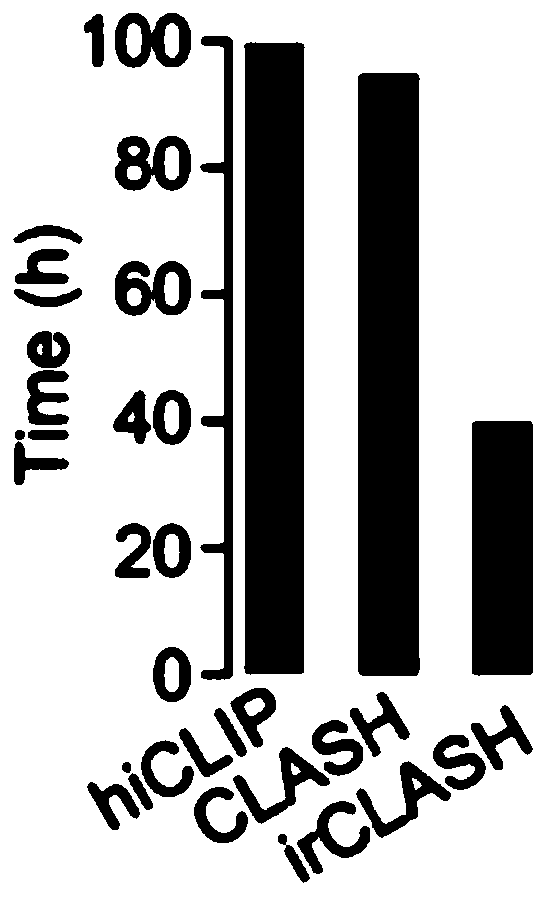
[0042] The overall process of the experimental method of the present invention is as follows: figure 1 As shown, the experimental sequencing library data analysis process is as follows figure 2 As shown, compared with other published methods, the results show that the method of the present invention greatly shortens the time-consuming experiments (such as image 3 ); the present embodiment takes the ADAR protein family ADAR1 protein as an example to further illustrate the present invention; specifically, the method includes the following steps:
[0043] 1. Cell preparation and protein and RNA cross-linking reaction
[0044] HEK293 cells were cultured in a 15cm cell culture dish and overexpressed FLAG-tagged ADAR1. After 48 hours, the cells were washed with PBS and the protein-RNA crosslinking reaction was performed on a 254nm ultraviolet cross...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com