Staphylococcus aureus rapid detection method based on enzyme digestion protection aptamer sensor
An aptamer sensor, staphylococcus technology, applied in biochemical equipment and methods, microbial determination/inspection, instruments, etc., can solve the problems of affecting the sensing accuracy and complicating the detection process, to simplify the operation steps, shorten the Simple effect of detection time and sample preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
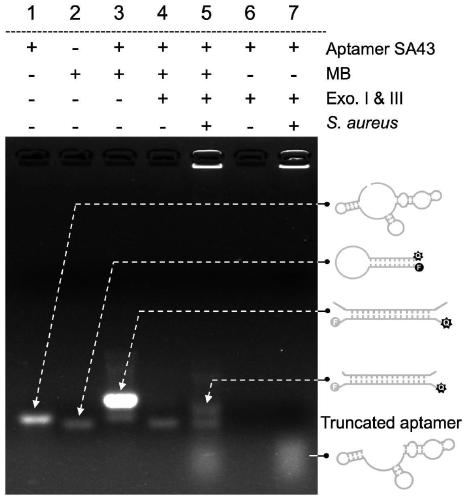
[0024] Example 1: Using Gel Electrophoresis Analysis to Verify the Working Principle of Enzyme Cut Protection Aptamer Sensor
[0025] The results of gel electrophoresis analysis were as figure 2 shown. It can be seen from the figure that the molecular beacon MB can be self-quenched when there is no aptamer SA43 (line 2, the fluorescence is dim); when MB is combined with SA43, the product band lags behind and the signal is significantly enhanced (line 3), indicating that SA43 Hybridized successfully with MB. When exonuclease I and exonuclease III were added, the band of SA43 disappeared (line4,6), indicating that the aptamer would be efficiently digested by exonuclease. And because the exonuclease has been inactivated before adding MB, Molecular Beacon MB will not be cleaved. When S. aureus is present, the strain can protect SA43 from complete exonuclease cleavage (line 5,7), but SA43 will be truncated because the band moves faster compared to the original aptamer (line 7)...
Embodiment 2
[0026] Embodiment 2: Optimization of reaction conditions
[0027] Take a certain amount of washed and resuspended Staphylococcus aureus bacteria liquid, mix with a certain volume and concentration of aptamer SA43, and react for 30 minutes, then add a certain concentration of exonuclease I and III to digest for 30 minutes, and then place it at 90°C After inactivating the enzyme activity for 5 minutes, add a certain amount of molecular beacon MB to the above mixture, mix well, place it at 90°C for 5 minutes, and measure the fluorescence signal after standing at room temperature for 10 minutes. The present invention optimizes the concentration of exonuclease I and exonuclease III, the concentration of molecular beacon MB, and the addition ratio of aptamer SA43 and MB. The optimization results were as follows: the amount of exonuclease I added was 0.16U / μL, the amount of exonuclease III added was 1.6U / μL, the ratio of SA43 to MB was 2:1, and the concentration of MB was 1 μM.
Embodiment 3
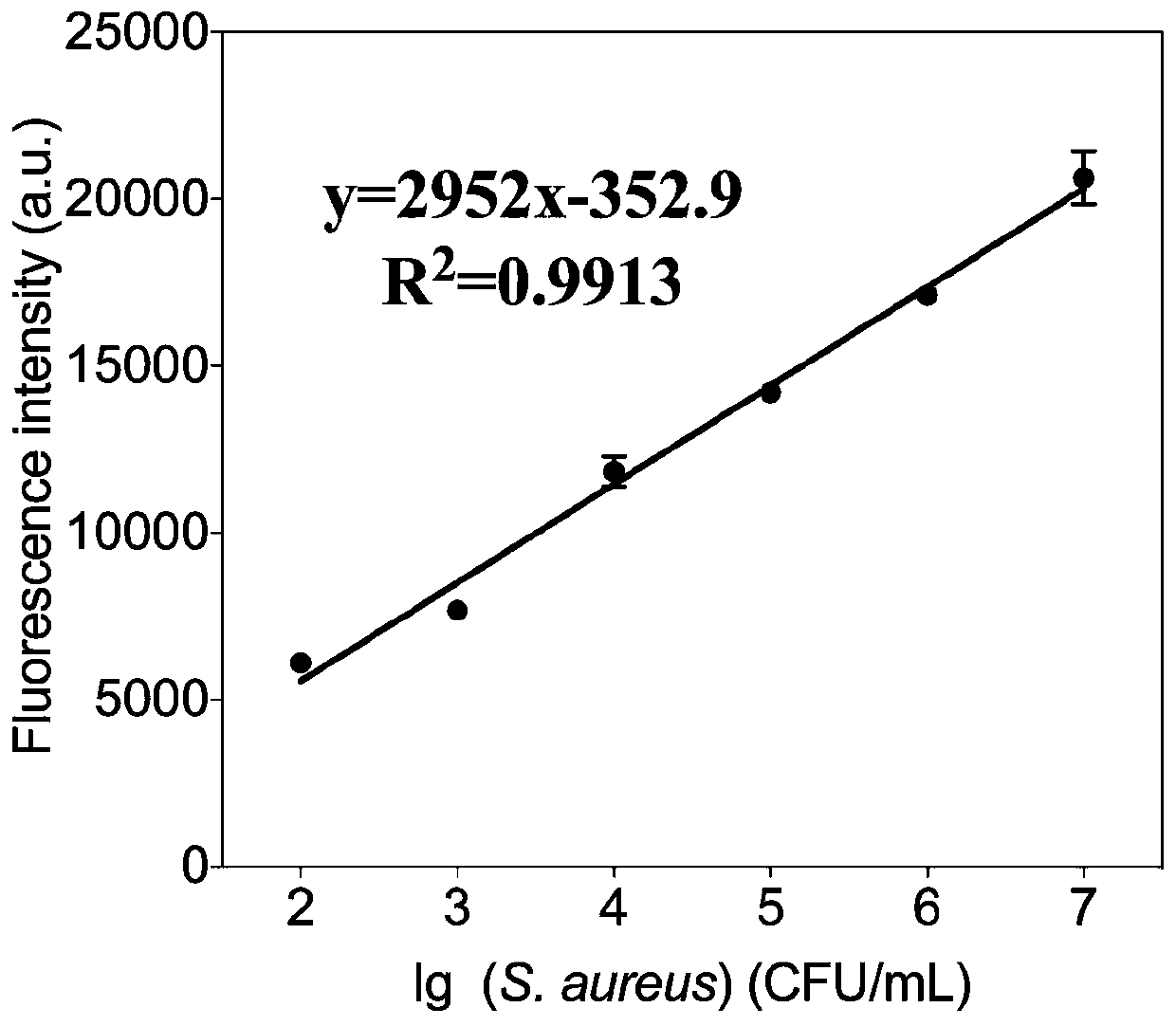
[0028] Embodiment 3: the drafting of Staphylococcus aureus standard curve
[0029] Preparation of Staphylococcus aureus suspensions with different concentration gradients: 0, 10, 10 2 ,10 3 ,10 4 ,10 5 ,10 6 ,10 7 ,10 8 ,10 9 ,10 10 ,10 11 CFU / mL, take 8 μL of the above bacterial suspension, mix with 8 μL of 20 μM aptamer SA43 solution, 8 μL of exonuclease I buffer (10x) and 46 μL of water, combine at 37°C for 30 min, then add 2 μL of exonuclease I Enzyme I (6.4U / μL) and exonuclease III (64U / μL) solution, mix well, place at 37°C for enzyme digestion for 30min, after digestion, place the mixture at 90°C for 5min to inactivate the enzyme, then Take 8 μL of 10 μM MB solution and add it to the above mixture, mix well, place it at 90°C for 5 minutes and let it stand at room temperature for 10 minutes, then measure the fluorescence signal. The experimental results are in 10 2 to 10 7 In the bacterial solution concentration in the interval, the fluorescence intensity and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com