Serum-based method for rapid identification of Brucella vaccine strain infection and field strain infection
A technology for Brucella and vaccine strains, which is applied in the field of protein spectrum detection, can solve the problems that have not been used to identify Brucella vaccine strain infection and natural wild strain infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Determination of characteristic proteins and construction of standard detection models
[0039] 1. Modeling training sample selection
[0040] The present invention adopts the serum sample of 30 routine Brucella wild strain infection patients, the serum sample of 30 routine Brucella vaccine strain infections, the vaccine strain of Brucella has S2 and A19, all Brucella vaccine strains Infected samples and Brucella wild strain infected samples were confirmed by epidemiological investigation and cysteine agglutination test. 20% of the sample size is used for model cross-validation.
[0041] 2. Enrichment of serum protein samples
[0042] Take out the magnetic bead kit (Bruker MB-WCX kit) from the refrigerator at 4°C, take out a tube of weak cationic magnetic bead suspension, turn it upside down manually, and mix the magnetic bead suspension completely for 1 minute; draw 60 μL of magnetic bead binding buffer ( BB) into a 200 μL sample tube, then add 10 μL of ...
Embodiment 2
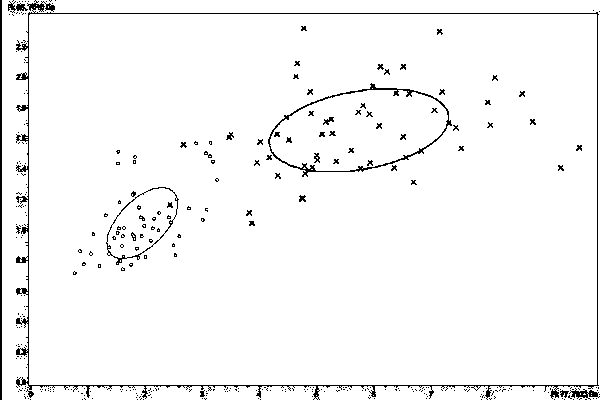
[0055] Example 2 Model Classification Ability Test
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com