Plant nitrilase mutant, coding gene and application of plant nitrilase mutant
A technology of nitrilase and coding gene, which is applied in the field of bioengineering, can solve the problems of inability to meet the needs of industrial production, low catalytic activity and stereoselectivity of nitrilase, and achieve the effect of improving catalytic activity and solubility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Construction of parental nitrilase gene
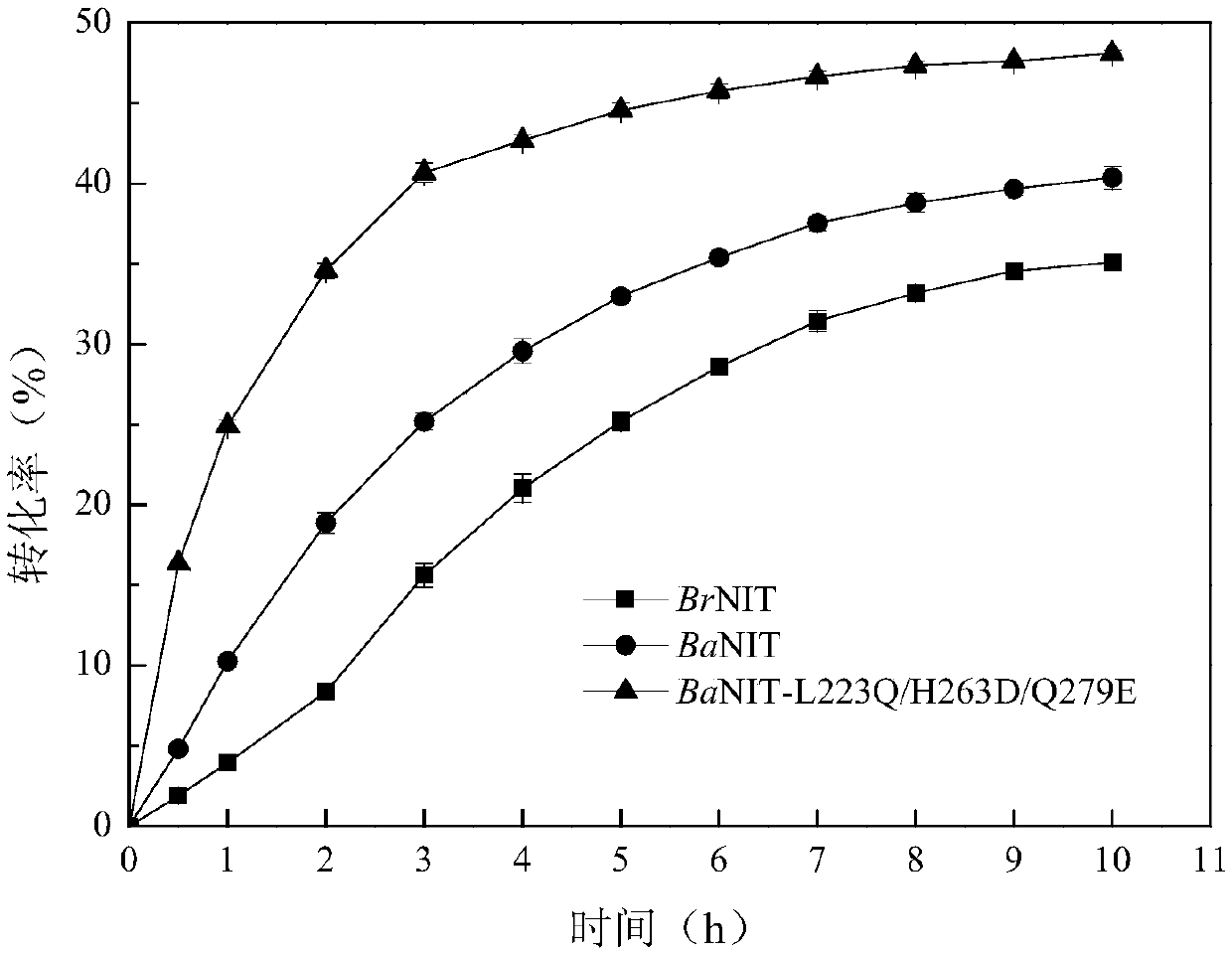
[0065] By comparing the nucleotide and amino acid sequences of cruciferous plant nitrilase, the key peptide 225-285 region was identified. See GenBank accession number: ABM55734.1 for the wild-type turnip nitrilase (BrNIT) sequence; see GenBank accession number: KFK44999 for the wild-type Alpine Arabidopsis thaliana nitrilase (AaNIT) sequence.
[0066] A one-step cloning method was used to insert the nucleotide sequence corresponding to the 225-285 peptides of Alpine Arabidopsis thaliana nitrilase (AaNIT) into the turnip nitrile hydrolase (BrNIT) plasmid fragment that lacked the corresponding peptide. Design primers are shown in Table 1.
[0067] Table 1: BaNIT chimeric enzyme primer design table
[0068]
[0069] Using the AaNIT nucleotide sequence as a template, clone the 225-285 peptide DNA fragment. PCR reaction system (50μL): Template DNA Master Mix, 0.2μM upstream and downstream primers, the rest ddH 2 O make up to the total vo...
Embodiment 2
[0076] Site-directed saturation mutations at positions 223, 263 and 279 of nitrilase
[0077] In order to mutate Leu(L) at position 223, His(H) at position 263 and Gln(Q) at position 279 in the parent amino acid sequence, corresponding primers were designed, as shown in Table 2. Show.
[0078] Table 2: Primer design table
[0079]
[0080] Note: N=A / G / C / T, K=G / T, M=A / C.
[0081] The recombinant plasmid pET28b-BaNIT containing the target gene fragment was used as the template, and the template was amplified by the whole plasmid according to the method of overlap extension PCR.
[0082] PCR amplification system (50μL): template DNA 0.1ng-1ng, 2×Phanta Max Buffer 25μL, dNTPs (10mM each) 1μL, mutation primer upstream and downstream each 1μL, Phanta Max Super-Fidelity DNA Polymerase 1U, the rest ddH 2 O make up to the total volume.
[0083] PCR reaction parameters: (1) 95°C pre-denaturation for 30s; (2) 95°C denaturation for 15s; (3) 63°C annealing for 15s; (4) 72°C extension for 6min, steps...
Embodiment 3
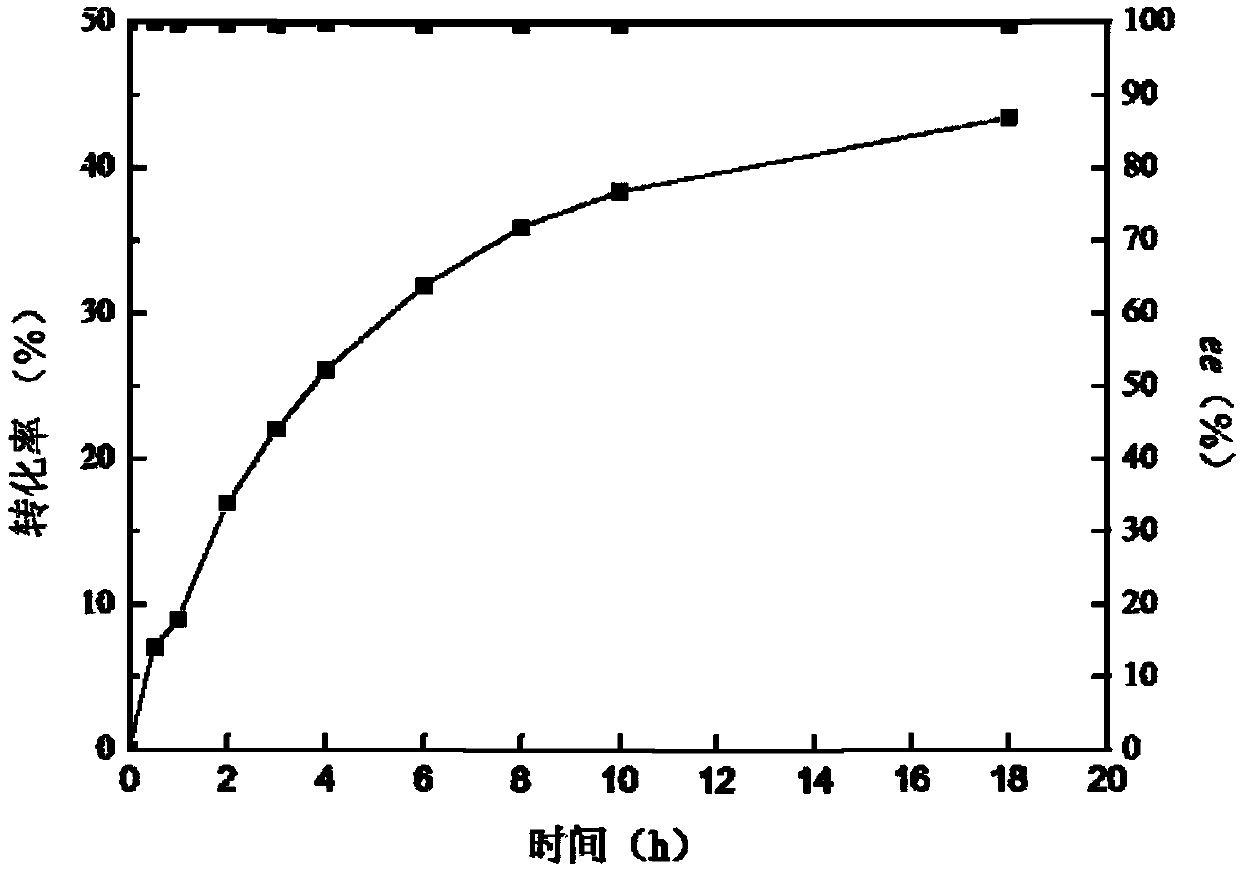
[0089] Construction of combined mutants of nitrilase
[0090] Using the expression plasmid pET28b-BaNIT-L223Q or pET28b-BaNIT-H263D as a template, site-directed mutagenesis was performed through whole plasmid amplification.
[0091] PCR amplification system (50μL): template DNA 0.1ng-1ng, 2×Phanta Max Buffer 25μL, dNTPs (10mM each) 1μL, mutation primer upstream and downstream each 1μL, Phanta Max Super-Fidelity DNA Polymerase 1μL, the rest ddH 2 O make up to the total volume.
[0092] PCR reaction parameters: (1) 95°C pre-denaturation for 30s; (2) 95°C denaturation for 15s; (3) 60°C annealing for 15s; (4) 72°C extension for 6min, steps (2)-(4) cycle 30 times; 5) Extend thoroughly at 72°C for 5 minutes and store at 4°C.
[0093] After the PCR product was positively analyzed by 0.9% agarose gel electrophoresis, take 20μL of PCR reaction solution, add 1μL of endonuclease Dpn I, digest the template plasmid DNA at 37℃ for 3h, and inactivate it at 65℃ for 10min. Transform into E.coli BL21(...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com