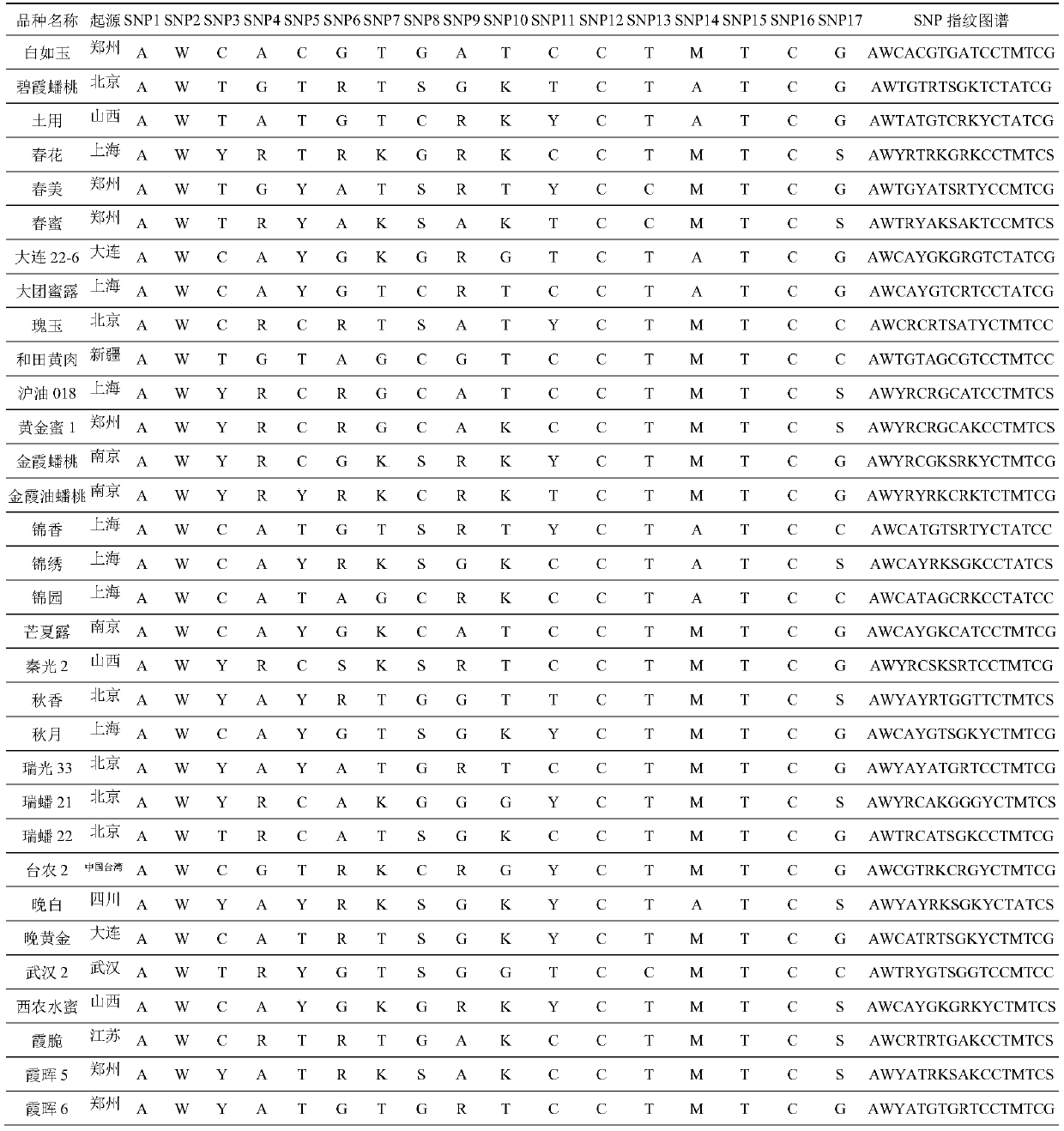
SNP molecular marker combination for constructing peach DNA fingerprint spectrum, application and method
A technology of DNA fingerprinting and molecular marking, applied in the direction of DNA / RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of technical stability, complexity and data integration not widely used
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Obtaining of SNP marker sites
[0057] In the present invention, 360 peach germplasms obtained from the peach germplasm resource nursery of Zhengzhou Pomology Research Institute, Chinese Academy of Agricultural Sciences are used as samples, and the sample DNA is extracted by the conventional CTAB method, and the 360 peach germplasms are resequenced by an Illumina HiSeq 2000 sequencer , to obtain 151Gb data, with an average sequencing depth of about 8.4×. According to the 50-150bp reads obtained by sequencing, compared with the peach reference genome version 2 (http: / / www.rosaceae.org / node / 355), 16658391 SNPs were identified.
Embodiment 2
[0058] Example 2 Filtering of SNP marker sites
[0059] Use the software vcftools v0.1.13 to filter the SNP marker sites, the specific method is as follows:
[0060] (1) Missing rate filtering
[0061] Use vcftools to calculate the deletion rate of each SNP site, filter out the SNP sites with deletions in 360 samples, and output a vcf file containing 939004 SNPs.
[0062] (2) SNP sequencing depth filtering
[0063] Calculate the sequencing depth of SNP in the vcf file that step (1) outputs, according to statistical result, filter out the SNP site that sequencing depth is lower than 10×, output the vcf file that comprises 310260 SNPs.
[0064] (3) SNP quality value filtering
[0065] Calculate the quality value Q of SNP in the vcf file that step (2) outputs, filter out the SNP locus with quality value lower than 1000, output the vcf file that comprises 141076 SNPs.
[0066] (4) π value filtering
[0067] Calculate the π value of SNP in the vcf file that step (3) outputs, f...
Embodiment 3
[0070] Example 3 Screening of SNP marker sites
[0071] (1) π value sorting
[0072] Carry out π value calculation to the vcf file containing 775 SNPs in step (5) of Example 2, output the txt file, sort the SNPs according to the π value, and use the SNP with the largest π value (Chr1: 9204153) as the first SNP of the set site.
[0073] (2) View sequence specificity
[0074] Use Samtools software to obtain the sequence of 150 bp upstream and downstream of the SNP (Chr1:9204153) molecular marker, and use blast (https: / / www.rosaceae.org / tools / ncbi_blast) to check the sequence specificity (E≥e-157).
[0075] (3) Determine the best SNP combination
[0076] The first SNP molecular marker (Chr1: 9204153) was combined with the remaining 774 SNP molecular markers (Perl program), and the SNP molecular marker (Chr2: 28221961), as the second SNP site.
[0077] Combining the set of the above two SNP molecular markers with the remaining 773 SNP molecular markers (Perl program), the SNP...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com