Genomic microsatellite wide-area length distribution estimation method considering tumor purity factors
A technology of length distribution and microsatellite, which is applied in the field of data science and can solve problems such as calculation deviation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
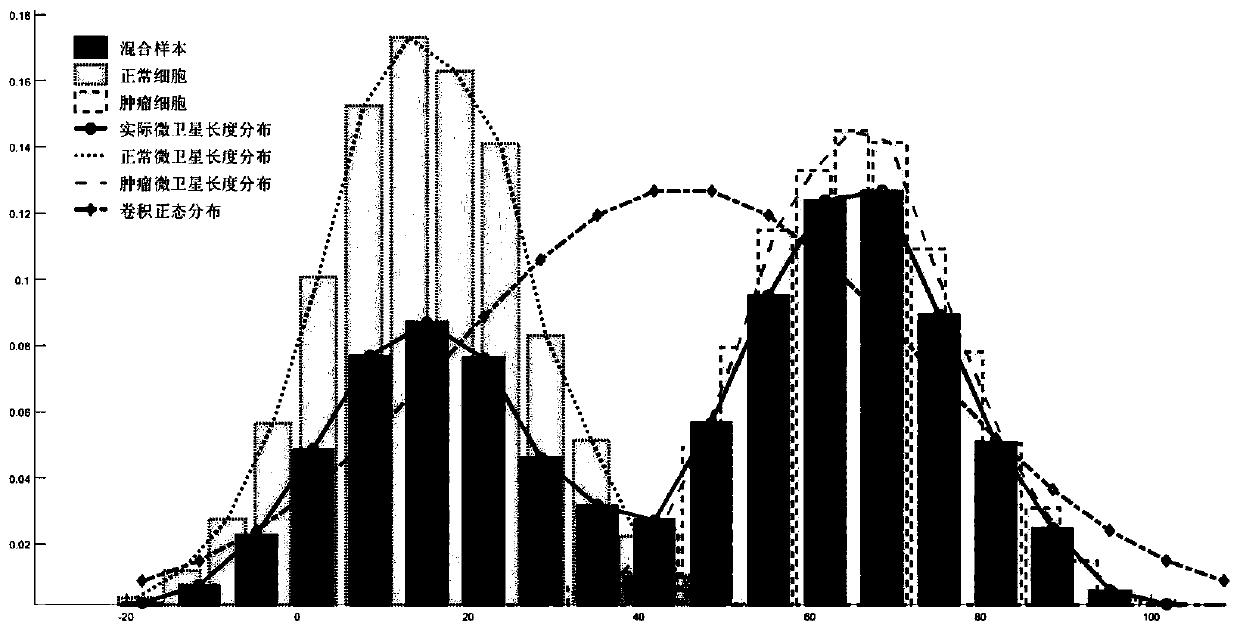
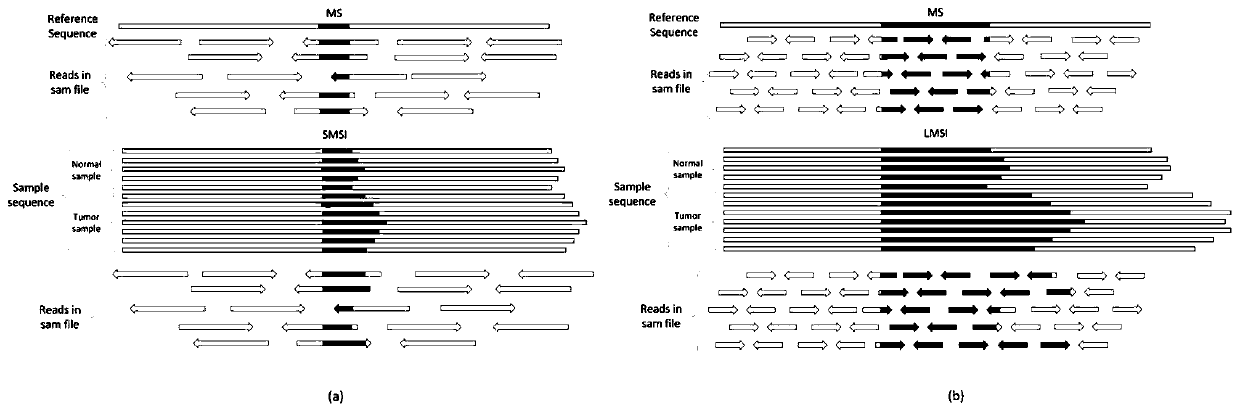
[0083] The present invention provides a genome microsatellite length distribution and state estimation method ELMSI (Estimation of Long Micro-satellite) based on tumor purity, the input data is the data of normal samples and their paired tumor samples, based on the purity estimation software, using The estimated tumor purity accelerates the process of deconvolution to solve the length distribution of mixed microsatellites, combining microsatellite read counting and maximum likelihood estimation to accurately identify the length distribution and status of short microsatellites in mixed samples, combined with maximum expectation Algorithms and the central limit theorem are used to infer the length distribution and state of long microsatellites, which solves the limitations of sample tumor purity and sequencing read length on MSI detection.
[0084] The present invention is based on following assumptions with general consensus in the academic circle:
[0085] 1. When the patient ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com