Method for dividing breeding swine blood linkage based on whole genome SNP information
A whole-genome and genome-wide technology, applied in the field of breeding pig pedigree based on whole-genome SNP information, can solve the problems of only considering the pedigree information from the father and the unreliable division method, and achieve a clear and concise interface, accurate division, and kinship Predict accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
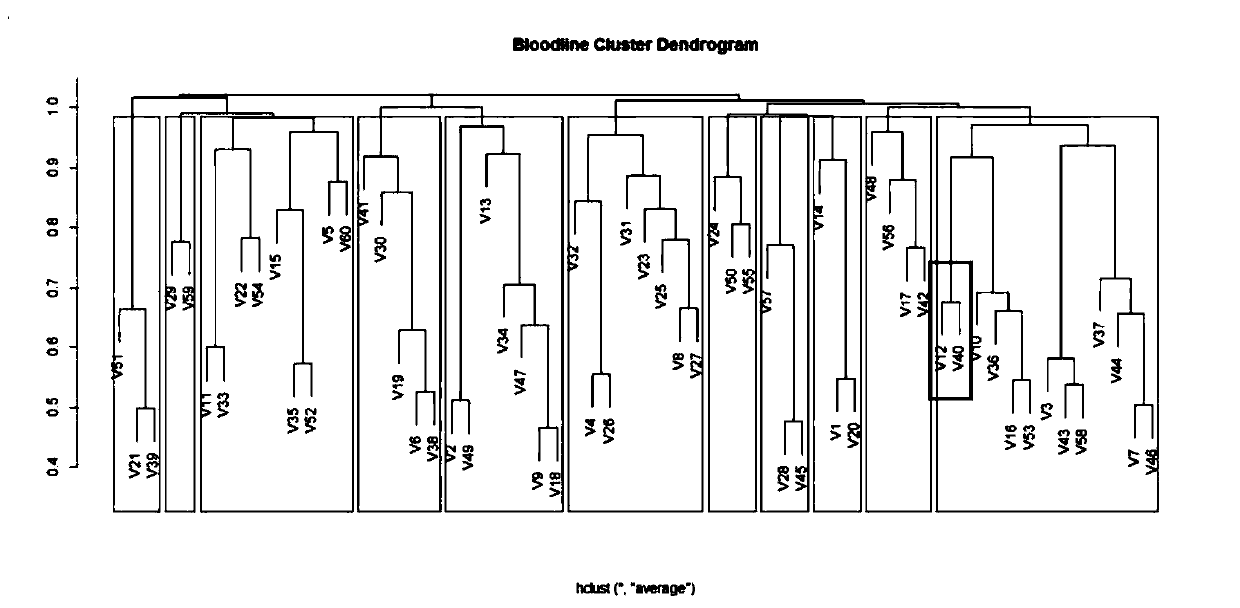
[0037] Example 1 Lineage division based on genomic SNP data
[0038] Experimental materials: The experimental materials are 60 breeding boars (the initial letter is represented by V) in the original breeding pig farm of Qingyuan No. 2 District, a subsidiary of Wen's Food Group Co., Ltd. All 60 breeding boars have been subjected to genomic SNP genotype mining by GBS (genotyping-by-sequencing) technology.
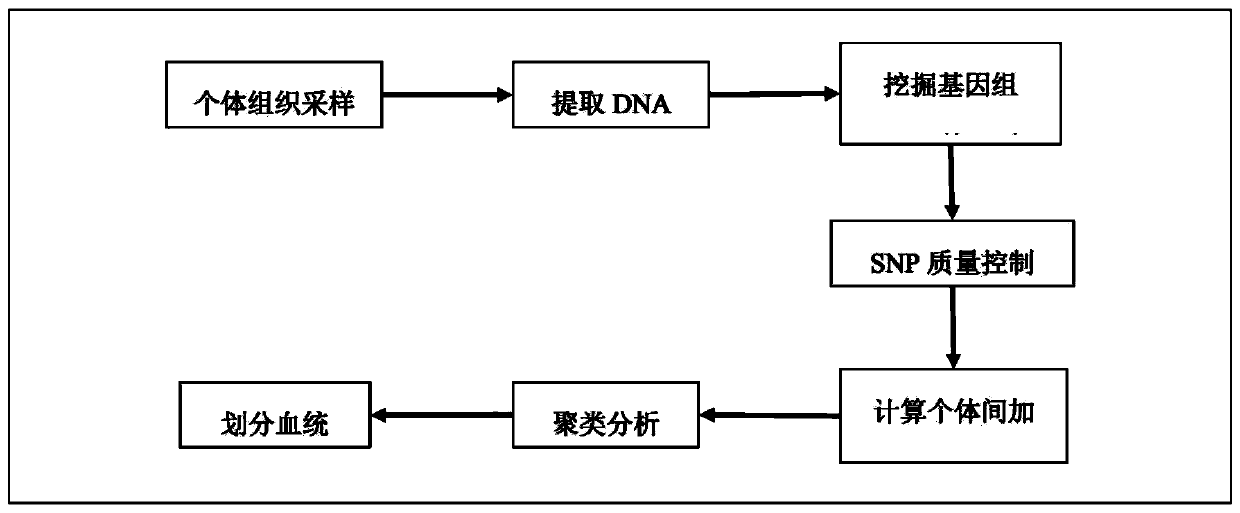
[0039] The flow chart of this embodiment is shown in figure 1 ,Specific steps are as follows:
[0040]DNA was extracted from ear tissues of all boars, and genomic libraries were constructed for all boars using the simplified genome (GBS) technology based on two restriction enzymes EcoRI and MspI, and sequenced on the Illumina HiSeq X Ten high-throughput sequencing platform. The obtained raw data were quality-controlled by Fastqc software, and the processed fragments were compared to the reference genome by BWA 0.7.15 software. After base quality correction, SNP typing was p...
Embodiment 2
[0042] Embodiment 2 new lineage division method compares with traditional division method
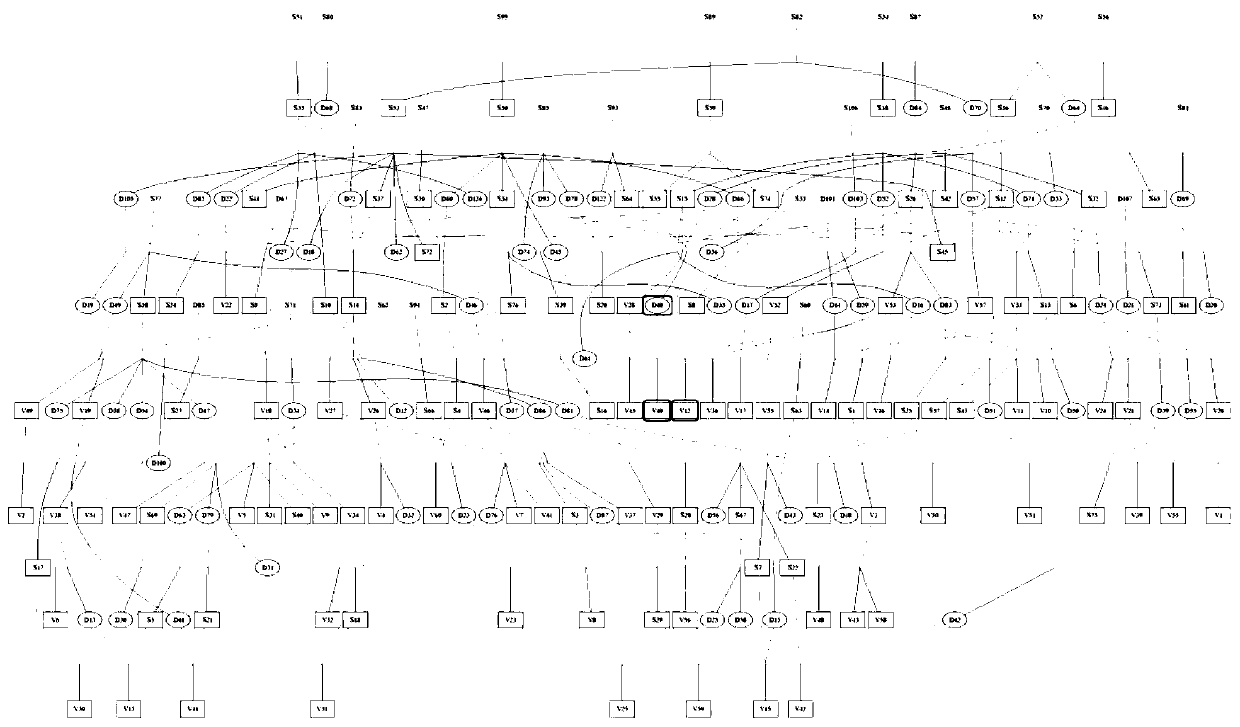
[0043] Experimental materials: All the ancestor information from the parents to the great-grandfather of the above 60 breeding boars was retrieved from the pedigree (except for the 60 boars, the initials of the boars are represented by S, and the sows are represented by D), all ancestors are not Genome SNP mining, as the material of traditional ancestry classification, is used to compare with the results of ancestry classification in the present invention.
[0044] Call out the pedigree of all 60 boars to the great-grandfather in the field, and draw the pedigree through Pedigraph software. After all the parents of the 60 boars are taken into account, the following is generated: image 3 From the results shown, it can be seen that the pedigree structure is extremely complex, and it is difficult to divide individuals into a certain lineage. Therefore, considering only absorbing informati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com