Fusion detection method for homologous genes based on differential SNP markers
A homologous gene and detection method technology, applied in the field of DNA sequencing, can solve the problems of undetectable signals, difficulty in determining the information of structural variation sites, and large influence of repetitive sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0085] The technical solutions of the present invention will be described in further detail below with reference to the accompanying drawings and embodiments.
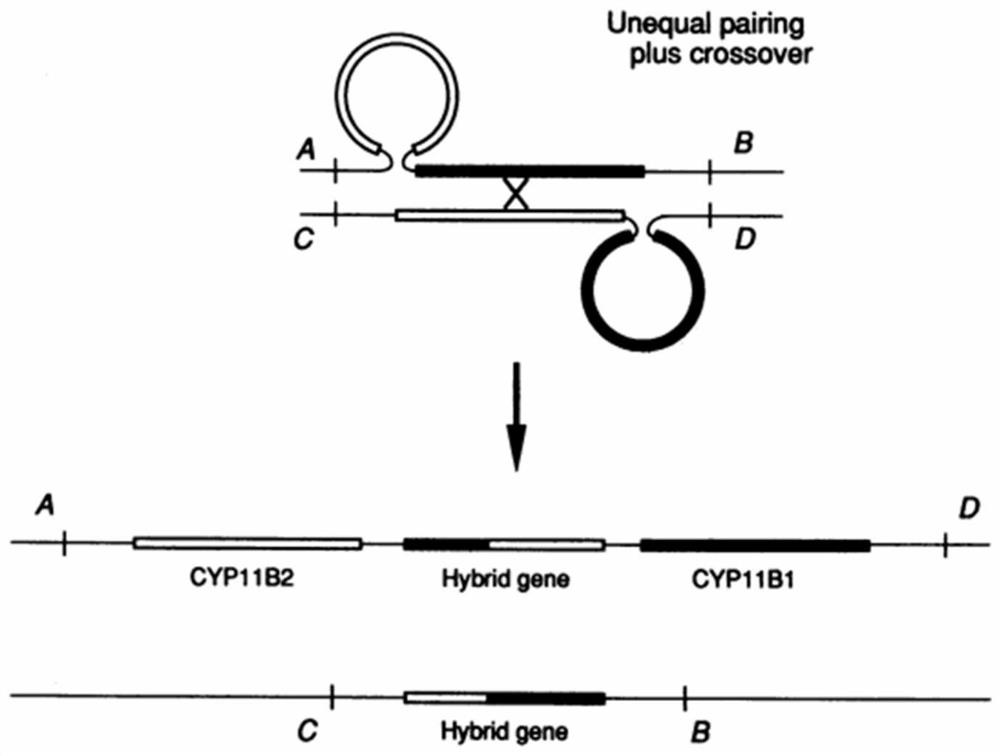
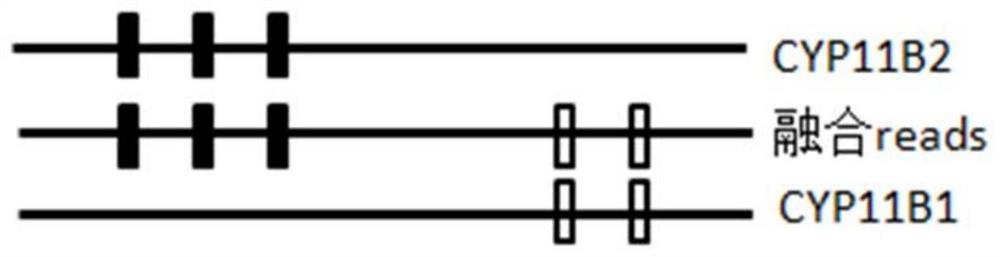
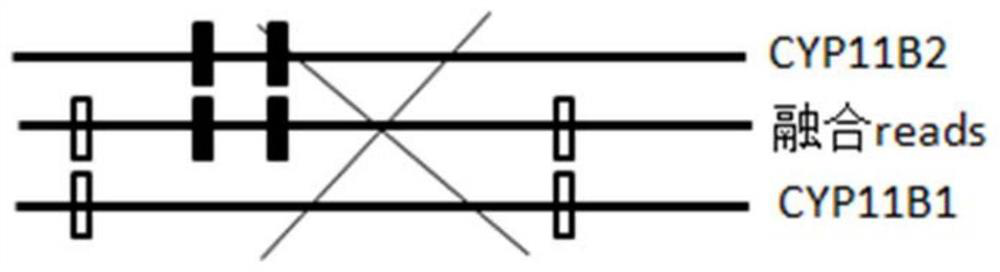
[0086] The present invention provides a method for judging the fusion of homologous genes based on differential SNP markers, such as Figure 1 to Figure 5 As shown, a preferred embodiment of the present invention is shown therein.
[0087] Specifically, the fusion determination method includes:
[0088] 1) Extract double-ended pair-end reads that meet the insert length conditions compared to the reference genome, and extract single-end reads that have SNP signals with the reference genome;
[0089] 2) Determine the SNP signal of double-ended pair-end reads or single-ended reads, compare the sequence consistency of each sequenced reads sequence with each homologous gene, find the continuous consistent SNP mark, obtain the breakpoint position, and Based on this, the region where the fusion is located is determined.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com