Method for treating biomolecules and method for analyzing biomolecules
一种生物分子、加热处理的技术,应用在生物分子的处理以及分析领域,能够解决纳米孔阻塞等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] A sample solution 1 was prepared by subjecting an aqueous solution having the following composition 1 to heat treatment and cooling treatment. Heat treatment and cooling treatment After maintaining at 95° C. for 15 minutes, it was rapidly cooled to 4° C. (cooling rate: 10° C. / second) and maintained at 4° C. for 5 minutes.
[0095] Composition 1: 4.0M tetramethylammonium salt, 0.1M Tris
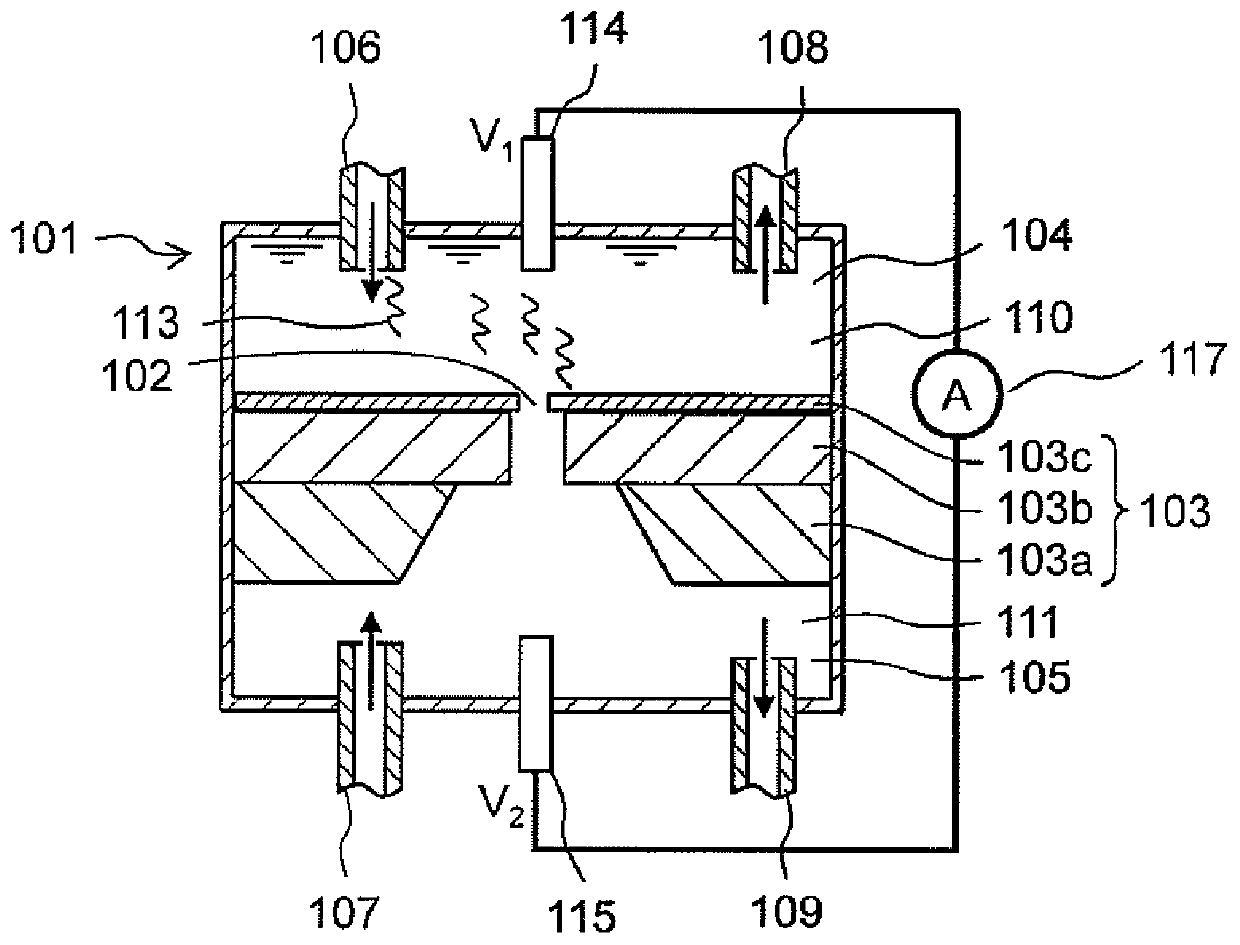
[0096] configured with figure 1 In the sample introduction region 104 of the nanopore analyzer having the structure shown, the diameter of the nanopore was 1.1 to 1.8 nm when the closed current generated when passing through the nanopore 102 was measured. In addition, the blocking current was detected using a patch clamp amplifier (Axopatch 200B amplifiers, manufactured by Molecular Devices, Inc.). The blocking current was detected under the conditions of a sampling rate of 50kHz and an applied voltage of +300mV. From the obtained detection data, evaluations were carried out for "blo...
Embodiment 2
[0098] A sample solution 2 was prepared by performing the heat treatment and cooling treatment described in Example 1 on an aqueous solution having the following composition 2. In addition, except that the sample solution 2 was used instead of the sample solution 1, the sealing current was measured and evaluated in the same manner as in Example 1.
[0099] Composition 2: 4.0M tetraethylammonium chloride, 0.1M Tris
Embodiment 3
[0122] A sample solution 9 was prepared by performing the heat treatment and cooling treatment described in Example 1 on an aqueous solution having the following composition 5. In addition, except that the sample solution 9 was used instead of the sample solution 1, the sealing current was measured and evaluated in the same manner as in Example 1.
[0123] Composition 5: 1.7M Potassium Chloride, 1.5M Tetraethylammonium Chloride, 0.1M Tris
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com