Space transcriptome detection chip and method
A detection chip and spatial transcription technology, applied in biochemical equipment and methods, bioreactor/fermenter combination, biochemical instruments, etc., can solve the problems of limited number of transcripts, lack of spatial information of tissue cells, etc., and achieve high resolution Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
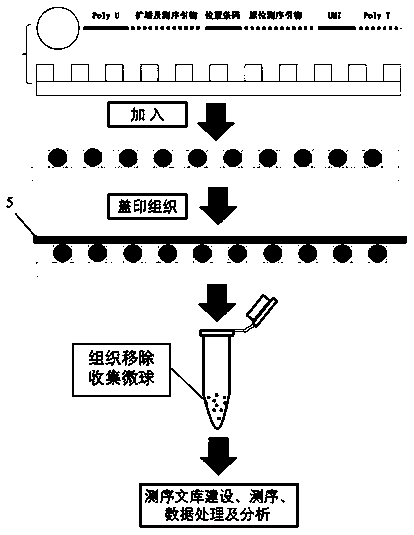
[0038] 1. Preparation of microplates
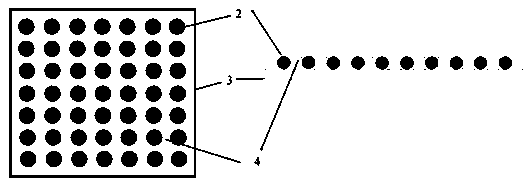
[0039] Design the size of the microwell plate 3 according to the experimental scale, the size of the well plate is 3 mm×3 mm, and microholes are etched on the silicon wafer as the initial mold, the depth of the microwells is 20 μm, the diameter of the microwells is 15 μm, and the spacing between the holes is 4 is 10 μm.
[0040] Pouring polydimethylsiloxane (PDMS) on the silicon wafer, and taking off the PDMS after molding, a reverse mold with micropillars can be made, and agarose prepared with enzyme-free water at a concentration of 5%, heated After melting, pouring is condensed on the reverse mold (PDMS microcolumn plate), and the agarose plate is peeled off to form a microporous plate 3 with a certain thickness. Add the DPBS-EDTA mixture that is harmless to the cells on the surface of the microwell plate 3 during storage, cover and store in a refrigerator at 4°C, and then the microwell plate 3 can be prepared, which can ensure that th...
Embodiment 2
[0065] Adult mice were euthanized and their brain tissues were taken, and tissue slices were prepared according to the methods in the examples.
[0066] All the other steps are the same as in Example 1.
[0067] After the construction of the gene sequencing library, the paired sequencing was performed on the Illumina NextSeq platform, and the gene expression profile was obtained after splitting, screening and comparison.
Embodiment 3
[0069] Adult mice were euthanized and their liver tissues were collected, and tissue sections were prepared according to the method in the examples.
[0070] All the other steps are the same as in Example 1.
[0071] After the construction of the gene sequencing library, the paired sequencing was performed on the Illumina NextSeq platform, and the gene expression profile was obtained after splitting, screening and comparison.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com