The sgRNA that specifically recognizes the porcine tert site, its coding DNA and its application
A specific identification and site technology, applied in the field of genetic engineering, can solve the problem that the molecular mechanism has not been fully analyzed, and achieve the effect of efficient large fragment knockout and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1 Design of sgRNA that specifically recognizes pig Tert site
[0020] According to the first exon sequence in the pig Tert gene sequence, its nucleotide sequence is as shown in SEQ ID NO:5 in the sequence table, design sgRNA; select the sgRNA target for gene knockout according to the PAM sequence, PAM sequence It is NGG, so the target of the sgRNA on the first exon sequence of the Tert gene is (named Tert-ag1): CGCCGCACGAAGGTGGCCAGCGG, and the nucleotide sequence identifying the target in the corresponding sgRNA sequence is shown in the sequence listing as SEQ ID As shown in NO: 1, the DNA sequence encoding the above sequence is shown in SEQ ID NO: 3 in the sequence listing.
[0021] According to the second exon sequence in the porcine Tert gene sequence, its nucleotide sequence is shown in SEQ ID NO: 6 in the sequence table, and the sgRNA is designed; the sgRNA target for gene knockout is selected according to the PAM sequence, and the PAM sequence It is NGG, ...
Embodiment 2
[0022] Example 2 Construction of sgRNA expression vector specifically recognizing pig Tert site
[0023] Purchase CRISPR / Cas9 rapid construction kits VK001-02 and VK001-04 from Weishang Lide Company, connect the Tert-sg1 reference kit instructions in the above Example 1 to the VK001-02 vector, and connect the Tert-sg1 in the above Example 1 -sg2 was connected to the VK001-04 vector according to the instructions, and the next experiment was carried out after it was verified to be correct by sequencing.
Embodiment 3
[0024] Example 3 Verification of the efficiency of sgRNA that specifically recognizes the porcine Tert site
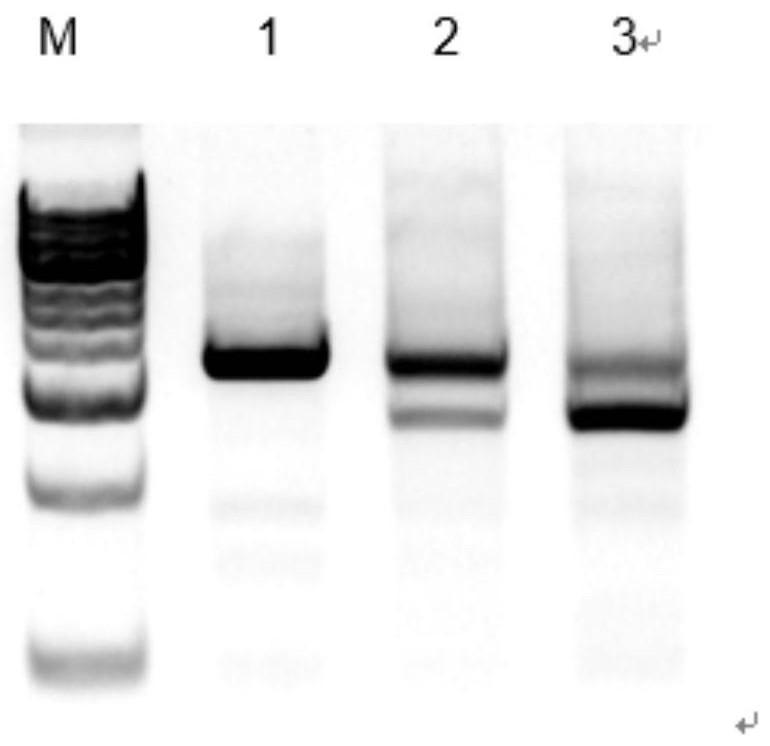
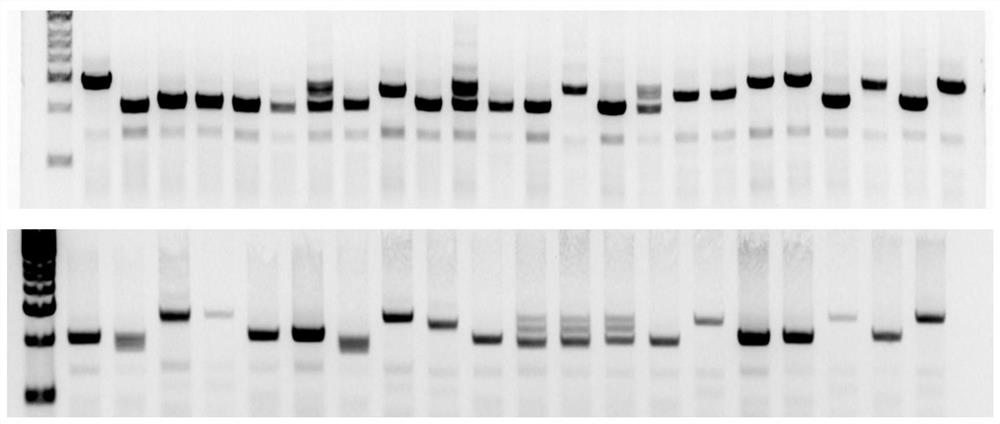
[0025] The Tert-sg1 expression vector and Tert-sg2 expression vector in the above Example 2 were co-transfected into porcine fetal fibroblasts by electroporation, and the cells were divided into two groups, one group was not enriched by flow cytometry, and the other was The group was enriched by flow cytometry, and the cells expressing both red fluorescence and green fluorescence were screened out. After 3 days of culture, the total DNA was extracted, and the Tert gene editing region was amplified. Templates, using the primers shown in Table 1, reacted with the system in Table 2, and the reaction conditions were 95°C for 5min; 95°C for 30s, 59°C for 30s, 72°C for 1min 30s 35×; 72°C for 5min.
[0026] Table 1 Primer sequence list
[0027]
[0028] Table 2 PCR reaction system
[0029]
[0030] The above reaction products were detected by electrophoresis, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com