Microsatellite instability detecting system and method based on genome sequencing
A genome sequencing and instability technology, applied in the field of microsatellite instability detection system based on genome sequencing, can solve the problems of poor stability of genome variation detection results, inability to detect, increase costs, etc., to reduce limitations and reduce costs , low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0054] The present invention will be described in detail below in conjunction with the accompanying drawings.
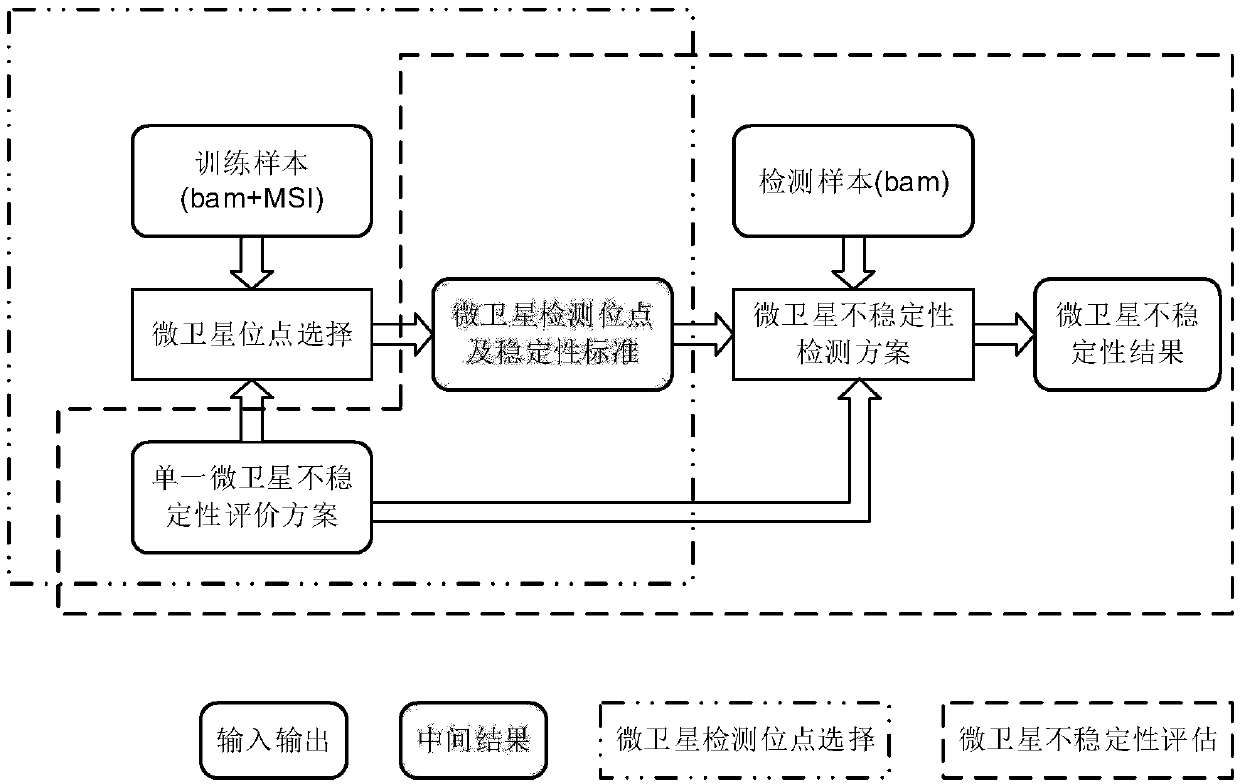
[0055] The invention provides a microsatellite instability detection system based on genome sequencing, including a microsatellite detection site selection module and a microsatellite instability detection module;
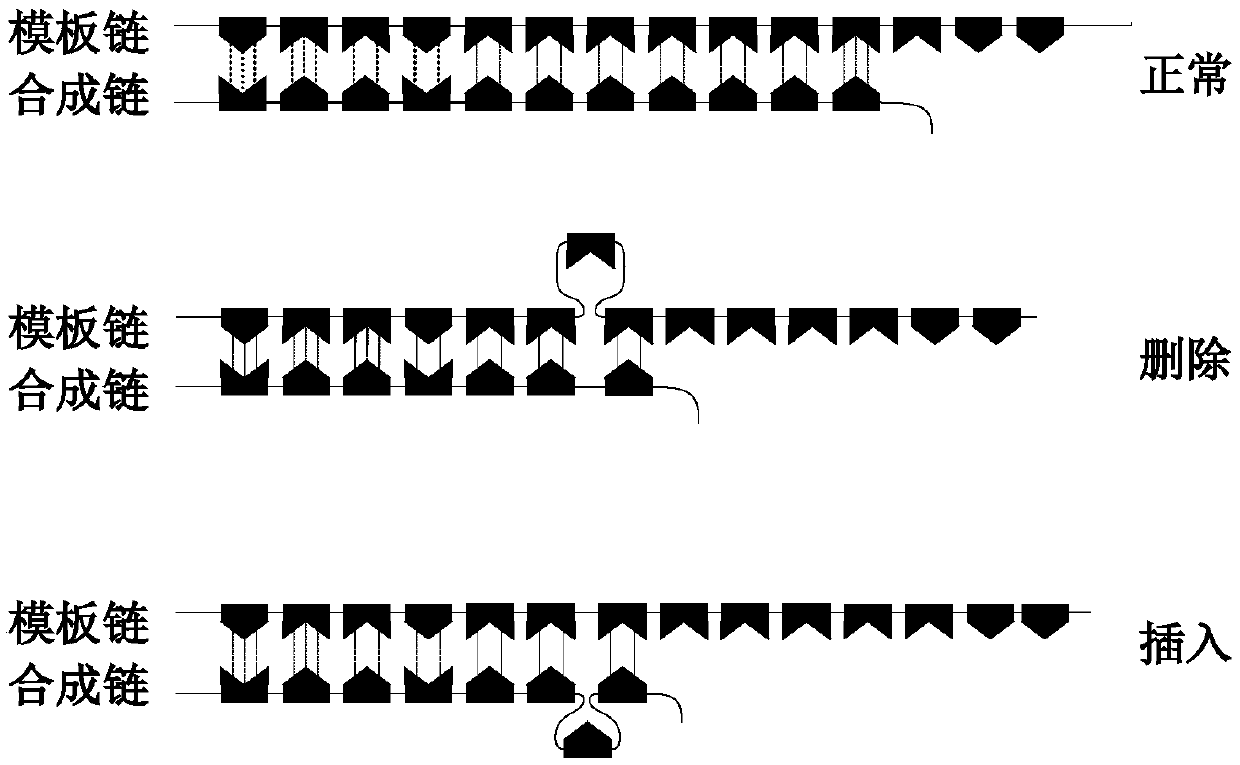
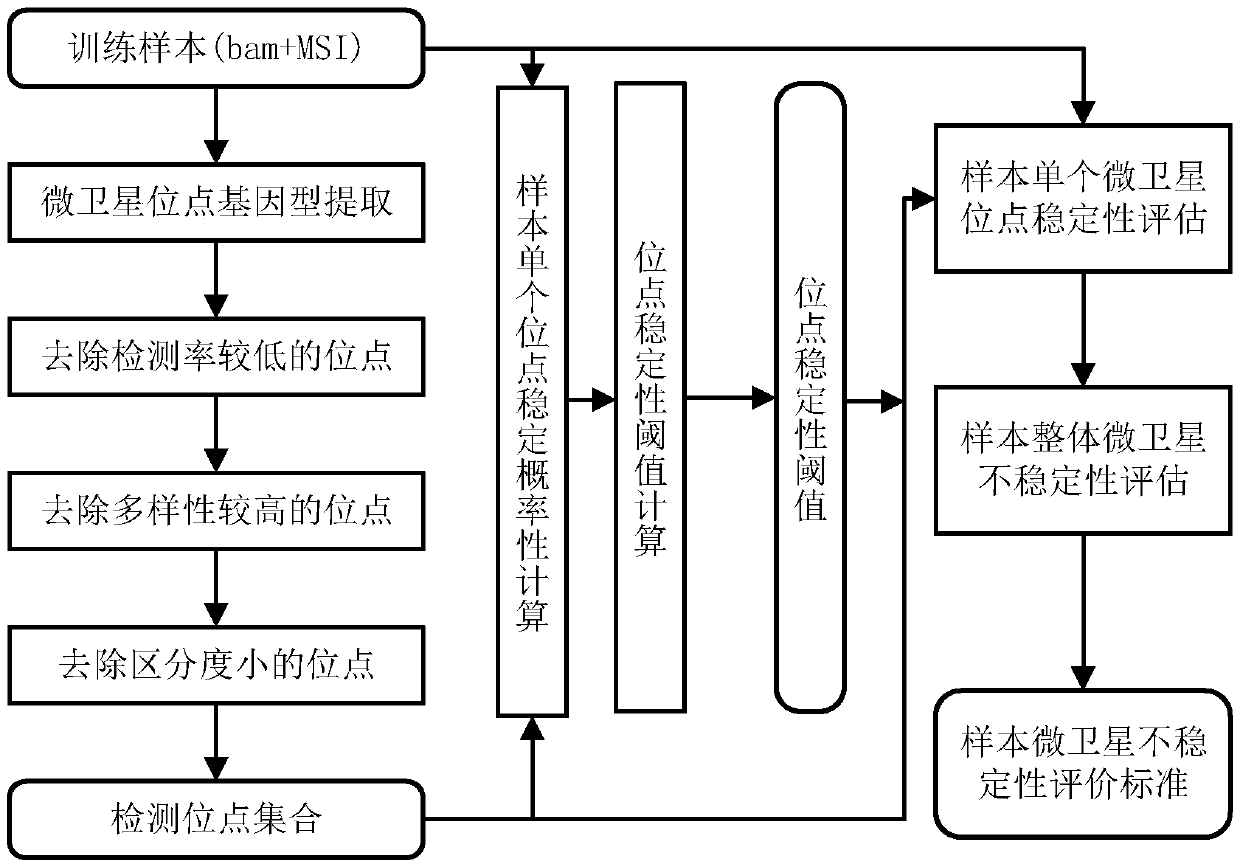
[0056] Wherein, the microsatellite detection site selection module is used to select an effective detection site based on the sequencing data of a certain tumor sample, and calculate the instability threshold of a single microsatellite site corresponding to the effective detection site and the threshold value of a certain tumor. The evaluation standard of sample microsatellite instability; the microsatellite instability detection module is used for the threshold value of the instability of a single microsatellite site corresponding to the effective detection site obtained by the microsatellite detection site selection module and a certain tumor sample The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com