Method for integrating multi-omics data to presume key protein kinases in cell transdifferentiation
A protein kinase and omics data technology, applied in the field of integrating multi-omics data to speculate key protein kinases in cell transdifferentiation, can solve problems that restrict the research and development of cell transdifferentiation mechanism, high cost of screening models, and lack of clear purpose , to achieve the effect of avoiding large-scale screening work, shortening the research and development cycle, and improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
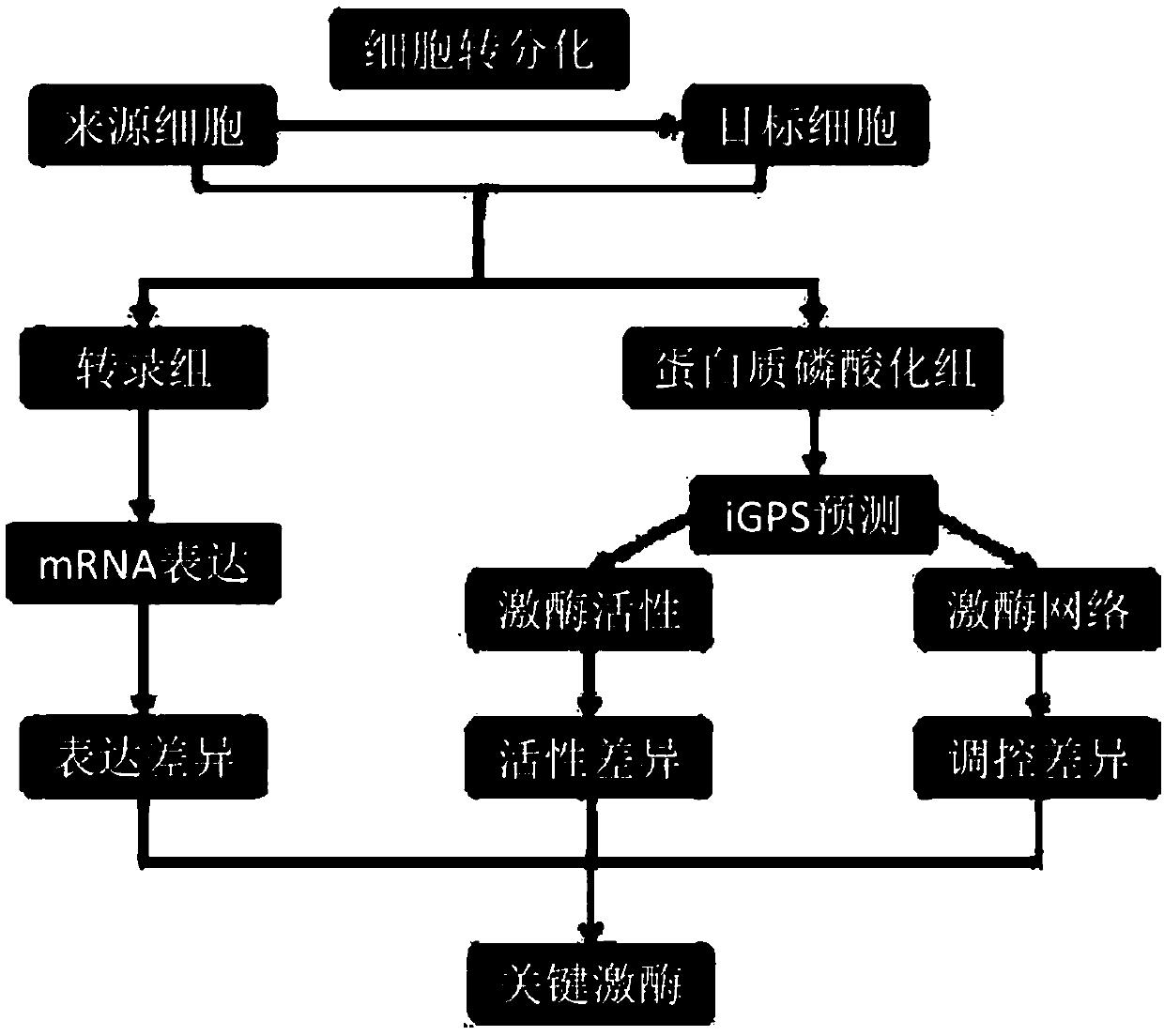
[0029] This example discloses in detail the method for inferring key protein kinases in cell transdifferentiation by integrating multi-omics data. The specific process is as follows: figure 1 As shown, this method is based on the data of omics detection, collects various cell samples in the process of cell differentiation, and through the significant differences in the expression, activity, and regulation of protein kinases in multiple samples, the cell transdifferentiation is obtained comprehensively. key protein kinases.
[0030] The detailed process is as follows:
[0031] (1) Prepare cell samples, divide cell samples evenly into transcriptome and protein phosphorylation group, conduct omics detection for each group, and obtain corresponding raw data;
[0032] Wherein, the cell samples include source cells that have not been transformed, transformed cells that have been successfully transformed, and initial state cells, intermediate state cells, mature state cells, and ste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com