Nucleic acid purification method for DNA methylation analysis of human stool
A technology of methylation and feces, applied in the field of obtaining nucleic acid samples for DNA methylation analysis, achieving a high conversion rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
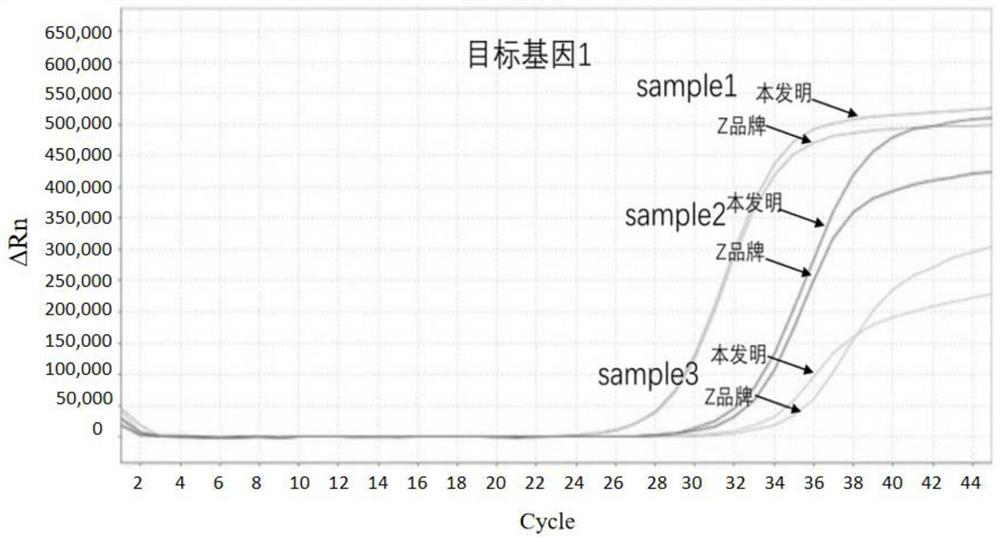
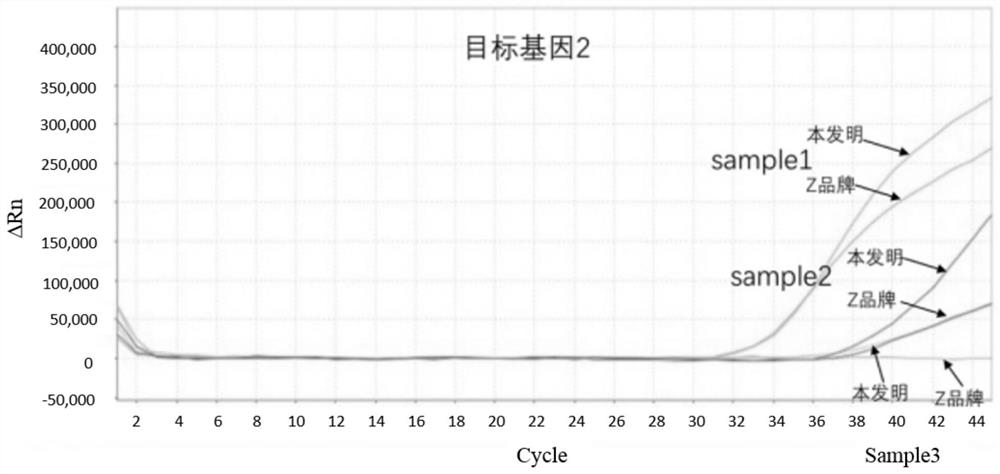
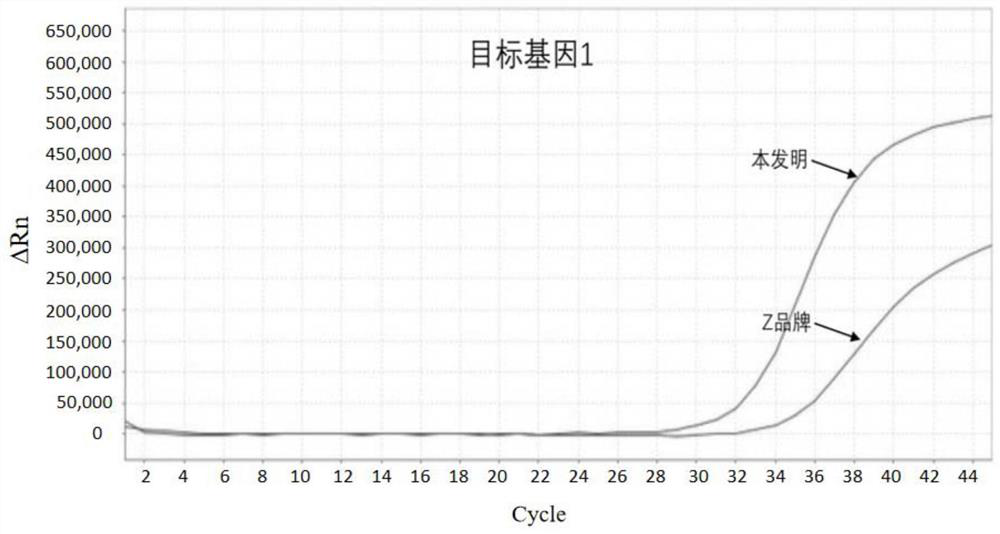
[0067] The method of the present invention of embodiment 1 human source feces total DNA is compared with Z brand commercialization sulfite conversion kit method
[0068] Acquisition of total DNA from human stool: select the stool samples of several patients with colorectal cancer, and obtain highly methylated DNA of the target gene through a nucleic acid extraction kit (produced by Shanghai Ruiyi Biotechnology Co., Ltd., No. 20180535). Human fecal total DNA.
[0069] One, the specific implementation steps of the nucleic acid purification method for DNA methylation analysis of the present invention:
[0070] (1) Add 2 μg of DNA to be processed (sample1, 2, 3) into a 2.0 mL centrifuge tube, and control the input volume range to 20-100 μL;
[0071] (2) After adding 190 μL of sulfite conversion solution and 30 μL of protective solution, cover the centrifuge tube tightly, mix it upside down, and centrifuge immediately;
[0072] (3) Seal the centrifuge tube and place it in a const...
Embodiment 2
[0109] The method of the present invention of embodiment 2 paraffin DNA is contrasted with Z brand commercialization sulfite conversion kit method
[0110] Paraffin DNA was extracted using a paraffin-embedded tissue DNA extraction kit ( DNA FFPE Tissue, 56404), to obtain FFPE-DNA.
[0111] One, the specific implementation steps of the nucleic acid purification method for DNA methylation analysis of the present invention:
[0112] (1) Add 500ng of FFPE-DNA to be treated in a 2.0mL centrifuge tube, the input volume is about 50μL;
[0113] (2) After adding 200 μL of sulfite conversion solution and 50 μL of protective solution, cover the centrifuge tube tightly, mix it upside down, and centrifuge immediately;
[0114] (3) Seal the centrifuge tube and place it in a constant temperature shaking incubator, and incubate at 95°C for 5 minutes; to prevent DNA renaturation, take out the centrifuge tube immediately and place it on ice for 5 minutes;
[0115] (4) Take out the centrifug...
Embodiment 3
[0152] The method of the present invention of embodiment 3 plasma free DNA is contrasted with Z brand commercialization sulfite conversion kit method
[0153] The plasma DNA was extracted using a free peripheral blood DNA extraction kit (VAHTS Serum / Plasma CircμLating DNA Kit, N902-01) to obtain cfDNA.
[0154] One, the specific implementation steps of the nucleic acid purification method for DNA methylation analysis of the present invention:
[0155] (1) Add 20ng of DNA to be treated into a 2.0mL centrifuge tube, with a volume of 50μL;
[0156] (2) After adding 280 μL of sulfite conversion solution and 50 μL of protective solution, cover the centrifuge tube tightly, mix it upside down, and centrifuge immediately;
[0157] (3) Seal the centrifuge tube and place it in a constant temperature shaking incubator, and incubate at 95°C for 5 minutes; to prevent DNA renaturation, take out the centrifuge tube immediately and place it on ice for 5 minutes;
[0158] (4) Take out the ce...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| percent by volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com