Single-cell Whole Genome Amplification and Library Construction Method for Improving Genome Coverage
A construction method and single-cell technology, applied in the field of molecular biology, can solve the problems of unsatisfactory single-cell whole-genome amplification methods, and achieve the effect of improving coverage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] (1) Single-cell gene amplification
[0061] A. Processing sorted cells:
[0062] Cells were sorted directly into 3 μL PBS in a clean bench without freeze-thawing.
[0063] B. Single-cell whole-genome amplification:
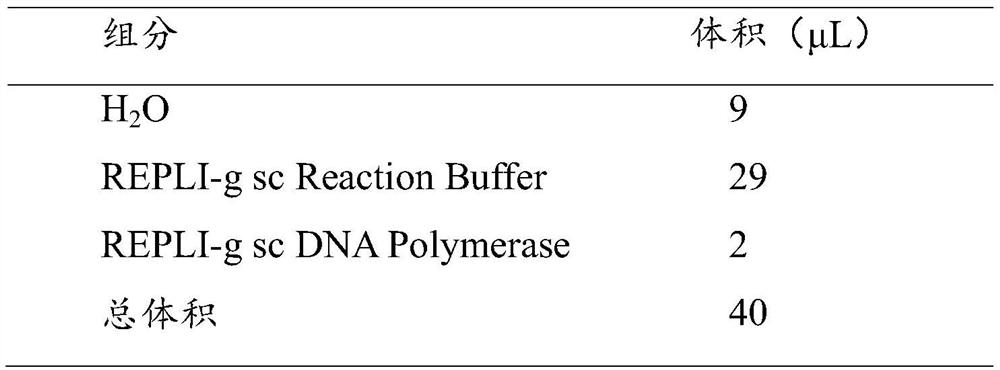
[0064] MDA was used for whole genome amplification, and Qiagen, REPLI-g Single Cell Kit was selected as the kit.
[0065] a) Prepare DLB buffer: Add 500 μL of water to the provided tube, mix thoroughly, and centrifuge briefly. DLBbuffer can be stored at -20°C for 6 months.
[0066] b) All buffers and reagents should be vortexed before use.
[0067] c) Adjust the temperature of the thermal lid of the PCR instrument to 70°C.
[0068] d) Prepare a sufficient amount of Buffer D2 (denatured buffer). Buffer D2 can be stored at -20°C for up to 3 months.
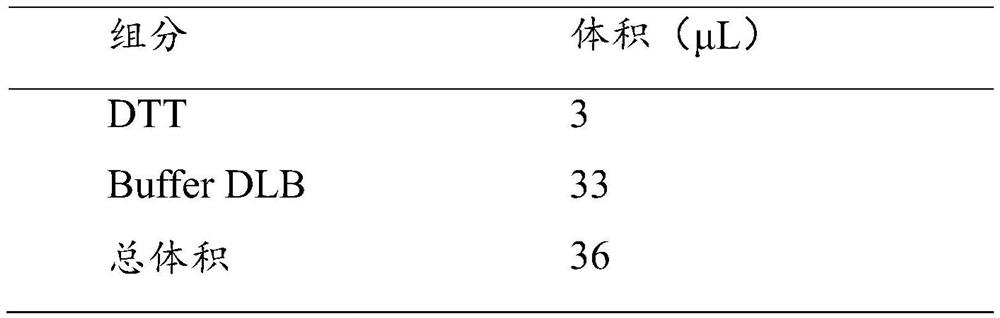
[0069] 3 μL of Buffer D2 is required for each reaction
[0070]
[0071] e) Make up the cell volume to 4 μL with PBS provided by the kit.
[0072] f) Add 3 μL Buffer D2, mix gently, and centrifuge brie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com