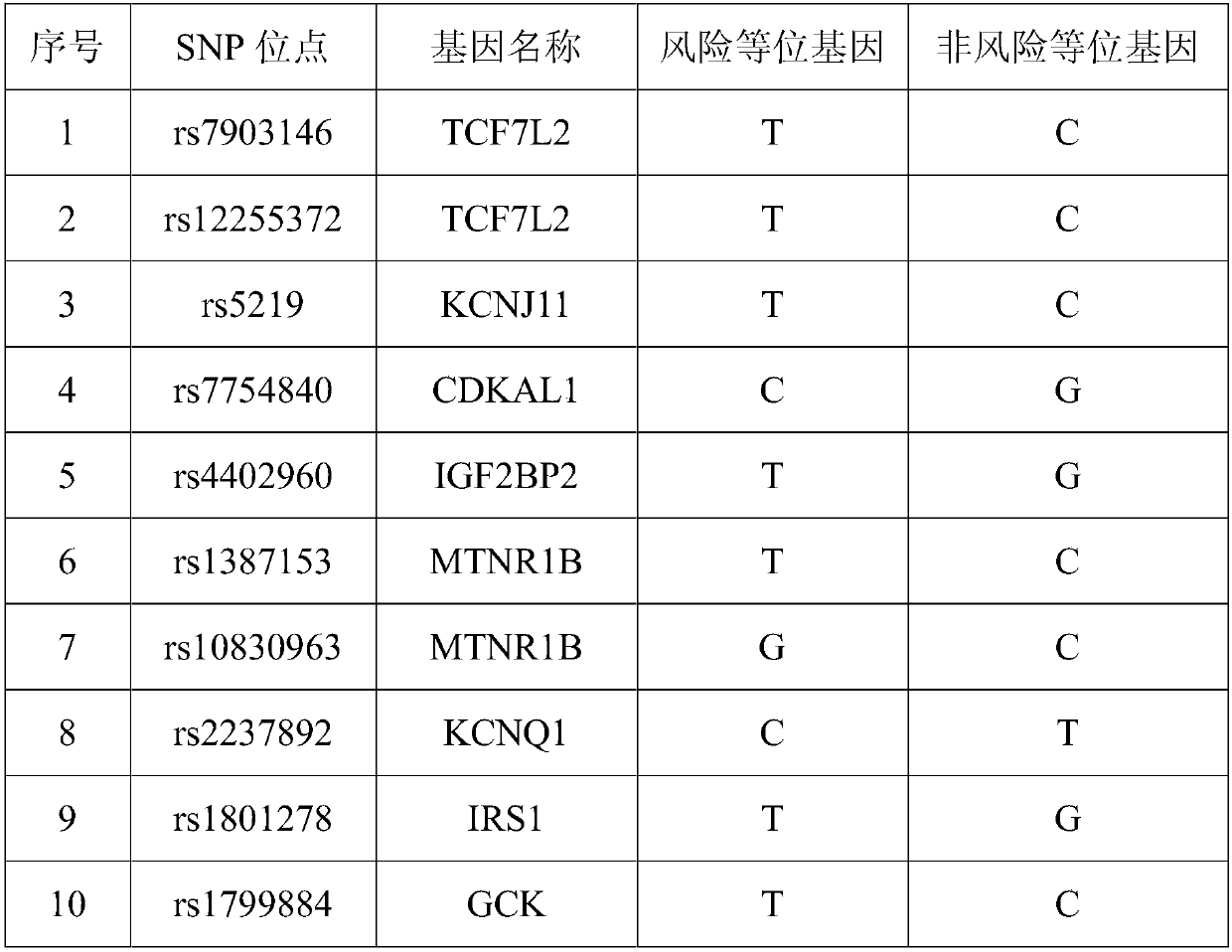
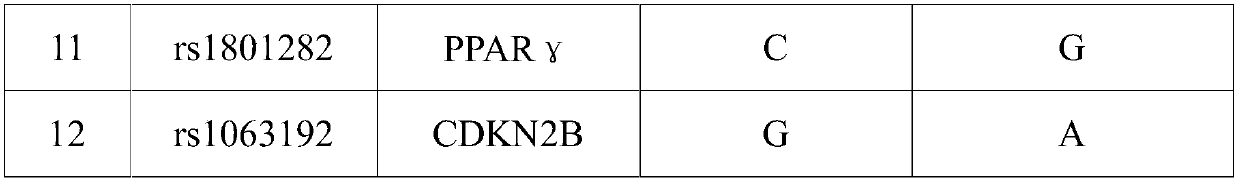
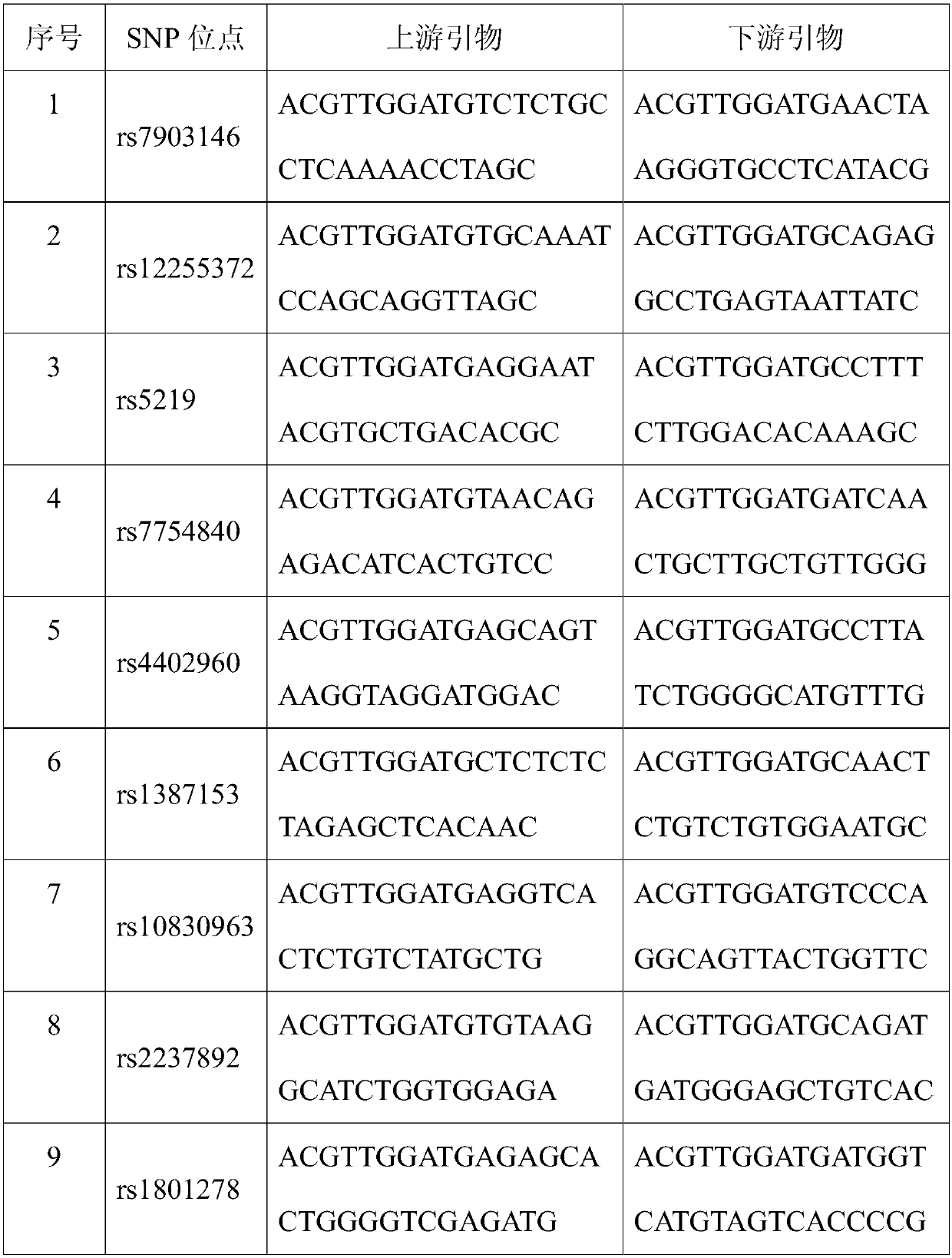
SNP (Single Nucleotide Polymorphism) marker for detecting diseasing risk of gestational diabetes and kit
A technology for gestational diabetes and risk of gestational diabetes, applied in the field of SNP markers and kits for detecting risk of gestational diabetes, to achieve the effect of solving early screening methods and high missed diagnosis rate, reducing prevalence rate, and lowering missed diagnosis rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment one: DNA extraction
[0040] 1.1 DNA extraction from saliva
[0041] Saliva DNA extraction was carried out using a buccal swab genomic DNA extraction kit (brand: Kangwei Century, item number: CW0530M).
[0042] Add a certain amount of absolute ethanol to Buffer GW1 and Buffer GW2. Take 500 μl saliva sample from the sampling tube, add 300 μl Buffer GR, 20 μl Proteinase K and 300 μl Buffer GL, shake and mix, incubate with shaking at 56°C for 15 minutes, add 300 μl absolute ethanol, shake and mix; add 750 μl of the mixed solution to the collection Add 400μl Buffer GW1 to the adsorption column, centrifuge at 12000rpm for 1min, discard the solution; add 400μl Buffer GW2 to the adsorption column, centrifuge at 12000rpm for 1min, discard the solution; centrifuge at 12000rpm for 2min, and Place to dry; put the adsorption column in a new 1.5ml centrifuge tube, add 40μl Buffer GE to the suspension, let it stand for 5min, centrifuge at 12000rpm for 1min, collect the D...
Embodiment 2
[0047] Embodiment two: PCR amplification
[0048] Add the DNA of the sample to be tested extracted in 1.1 or 1.2 of Example 1 into a 384-well plate for multiplex PCR amplification. Add the reaction system (total 5 μl) according to multiplex PCR amplification into each reaction well, the reaction system:
[0049] Template DNA: 1 μl,
[0050] Primer mix: 1 μl,
[0051] 10× PCR buffer: 0.5 μl,
[0052] 25mM MgCl 2 : 0.4 μl,
[0053] 25mM dNTPs: 0.1 μl,
[0054] wxya 2 O 1.8 μL,
[0055] The balance is Hotstar Taq enzyme (5U / μl).
[0056] After the reaction system is prepared, seal it tightly with PCR parafilm to prevent the sample from evaporating, shake and mix, and centrifuge; place the sealed 384-well plate on the ABI9700 PCR instrument for the following reactions:
[0057] Pre-denaturation at 95°C for 2 minutes;
[0058] 95°C for 30s, 56°C for 30s, 72°C for 1min, 45 cycles;
[0059] 5min at 72°C, keep at 4°C;
[0060] The PCR amplification product was obtained and...
Embodiment 3
[0061] Embodiment three: SAP digestion reaction
[0062] Using the matching reagents and operating steps of the Sequenom platform, the SNP site typing results of the samples to be tested are completed. The PCR amplification product obtained in Example 2 was added to each reaction well according to the reaction system (2 μl) digested by SAP, the reaction system:
[0063] SAP×Buffer: 0.17μl,
[0064] SAP Enzyme (1U / μl): 0.3μl,
[0065] wxya 2 O: 1.53 μl;
[0066] After the reaction system is prepared, seal it tightly with PCR parafilm to prevent the sample from evaporating, shake and mix, and centrifuge; place the sealed 384-well plate on the ABI9700 PCR instrument for the following reactions:
[0067] 37°C, 40min; 85°C, 5min; 4°C hold;
[0068] The PCR product after treatment with alkaline phosphorylase was obtained and centrifuged for future use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com