Method and device for detecting single sample body cell mutation sites in abnormal tissue and storage medium
A somatic cell mutation and mutation site technology, which is applied to the detection method, device and storage medium of a single sample of somatic cell mutation site in abnormal tissues, can solve the problem of not being able to contain reproductive genetic mutation information, and achieve high sensitivity and high specificity sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
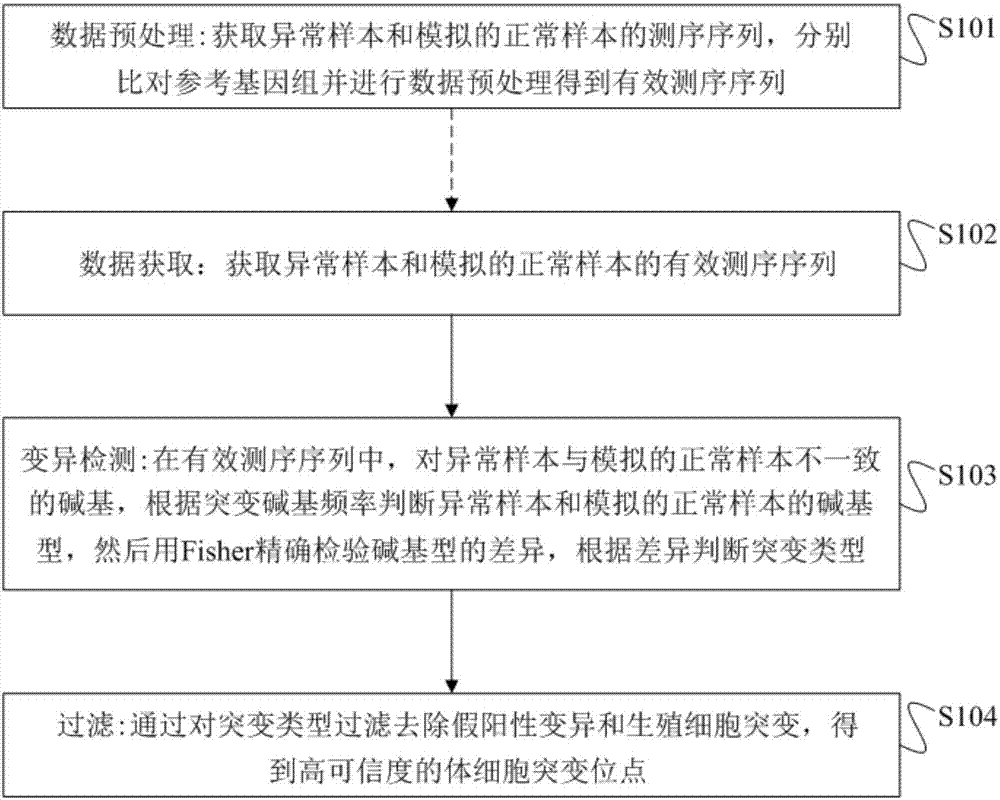
[0110] For 26 exome tumor samples with a sequencing depth of 100X or more, the single-sample somatic mutation site detection method of the embodiment of the present invention was used for somatic cell mutation detection, and the detection method based on paired samples was used for somatic cell mutation detection.
[0111] In this embodiment, the single-sample somatic mutation site detection method, specific methods and parameters include:
[0112] Data preprocessing: the abnormal sample is a tumor sample; the simulated normal sample is to extract more than a dozen original sequences obtained by next-generation sequencing and combine them into a 100X deep sample; the reference genome is a human reference genome; the filtering of effective sequencing sequences includes: Filter out non-human genome sequences and sequencing repeats; filter out sequences whose sequence comparison quality value is less than 1; for paired sequences in the overlapping region: if two paired sequences h...
Embodiment 2
[0124] According to the specific method and parameters of Example 1, the mutation of the important gene (CGC) of the tumor sample is detected by the single-sample somatic mutation site detection method of the embodiment of the present invention, and the mutation of the important gene (CGC) is analyzed Get back the situation.
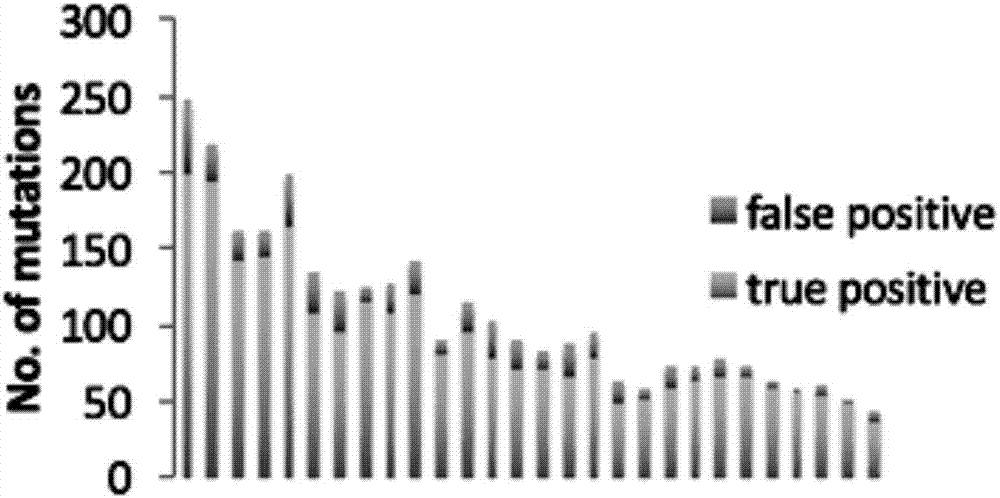
[0125] Such as Image 6 As shown, the column represents the number of CGC gene mutations (No.of mutations in CGC genes) of true positive and true negative (true negative) of the sample, showing that more than 90% of the mutations can be retrieved, and the TP53 and NOTCH1 genes are in The frequencies of mutations in esophageal squamous cell carcinoma of the test data are 85% and 12%, respectively, and the single-sample somatic mutation site detection method in the embodiment of the present invention can find 100%.
[0126] Such as Figure 7 As shown, one of the samples T99, the mutation retrieved using the single-sample somatic mutation site detection m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com