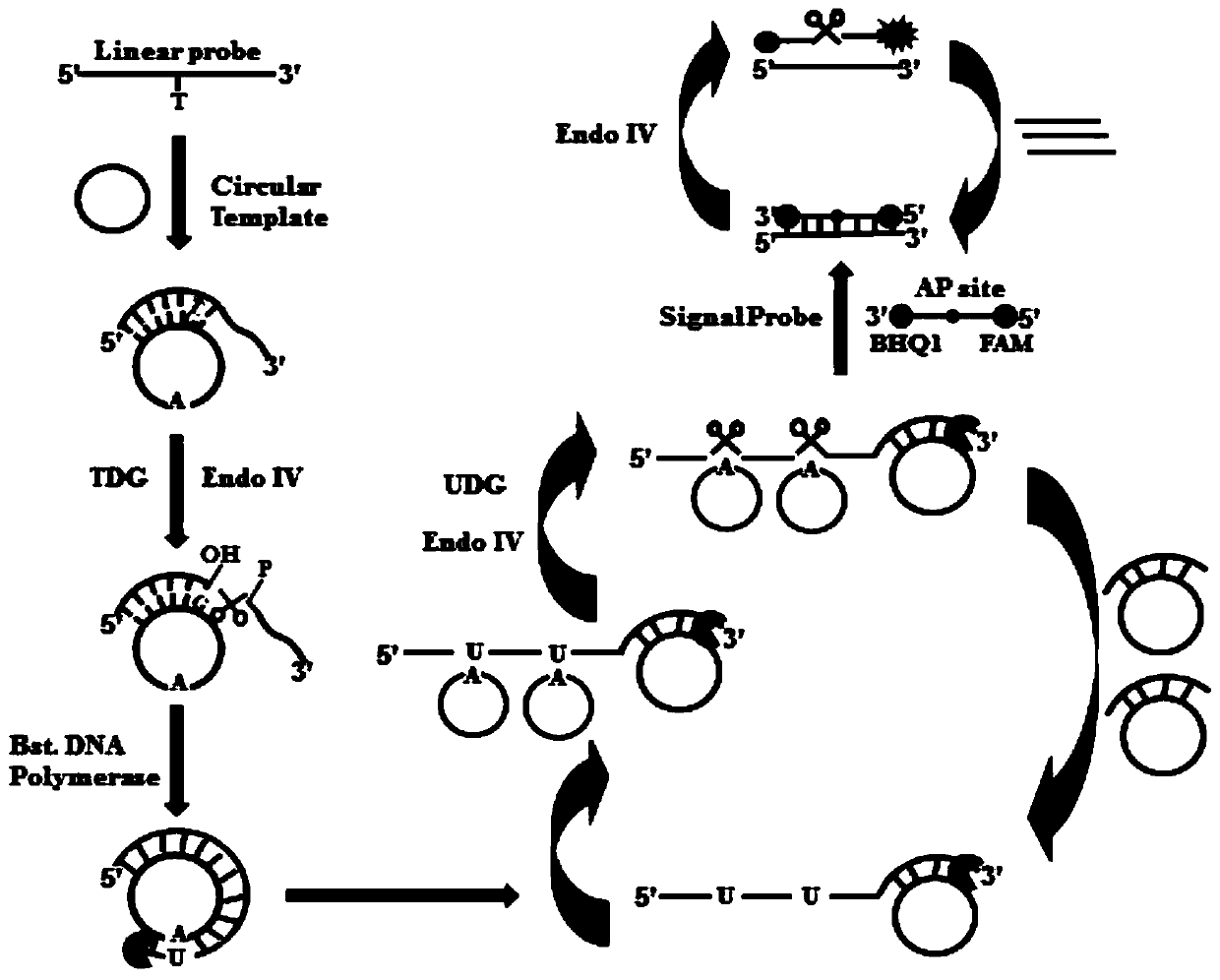
A method for the detection of thymidine-DNA glycosylase activity based on a dual-signal amplification strategy mediated by cyclic enzyme repair
A double-signal amplification and thymine technology, applied in the field of biological analysis, can solve the problem of limited sensitivity improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Preparation of circular template DNA: Dilute the ligation probe and linear padlock probe with 1× Tris(hydroxymethyl)aminomethane-ethylenediaminetetraacetic acid (Tris-EDTA) buffer to 10 micromoles per liter, and in Denature at 95°C for 5 minutes. Then 2 microliters of ligation probe and linear padlock probe were added to 20 microliters of ligation buffer, including 1×T4 ligase buffer (6.6 mmol per liter of magnesium chloride, 10 mmol per liter of Dithiothreitol, 0.1 mmol per liter of adenosine triphosphate, 66 mmol per liter of tris(hydroxymethyl)aminomethane-hydrochloric acid (Tris-HCl) (pH 7.6)), 50 units of T4DNA ligase, in Incubate overnight at 16°C. After the ligation reaction, transfer 10 µl of the ligation product to 10 µl of digestion buffer, including 1 mmol / L of dithiothreitol, 6.7 mmol / L of MgCl, 67 mmol / L of glycine - Potassium hydroxide (pH 9.5), 10 units of exonuclease I and 20 units of exonuclease III, incubated at 37°C for 2 hours, then inactivated at ...
Embodiment 2
[0063] Cell lysis buffer preparation: 10 mmol per liter of tris(hydroxymethyl)aminomethane-hydrochloric acid (Tris-HCl) (pH8.0), 150 mmol per liter of sodium chloride, 1% (mass / volume) Ethylphenyl polyethylene glycol (NP-40), 0.25 mmol per liter of sodium deoxycholate, 1% (mass / volume) glycerol, 0.1 mmol per liter of 4-(2-aminoethyl ) Benzenesulfonyl fluoride hydrochloride.
[0064] Cell extract preparation: Human cervical cancer cell (HeLa) and human breast cancer cell (MCF-7) culture medium is Dulbecco's modified Eagle's-Egg with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin Dill medium (DMEM), placed in an incubator containing 5% carbon dioxide, 37 degrees Celsius for cultivation. When the cells grow to the logarithmic growth phase, they are digested with trypsin and washed with ice phosphate buffer (137 mmol sodium chloride solution, 2.7 mmol potassium chloride solution, 10 mmol phosphate buffer saline, pH 7.4) Wash twice, then centrifuge at 4°C and 800 rpm...
Embodiment 3
[0070] 3.1 Experimental verification of the principle
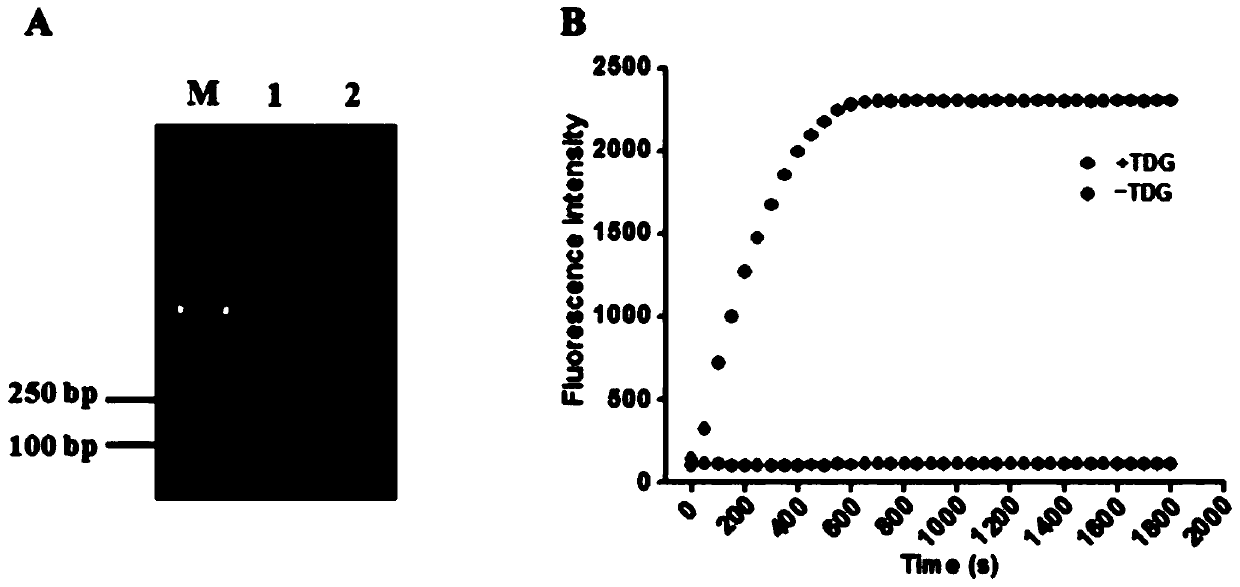
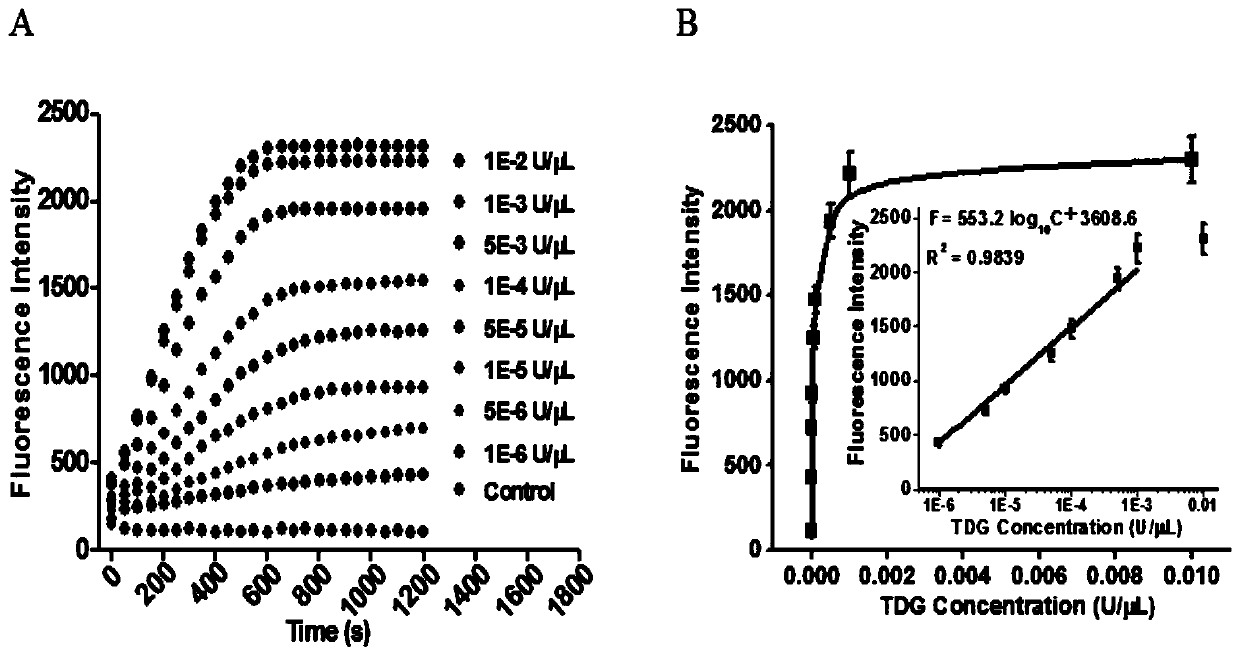
[0071] In order to verify the feasibility of this scheme, we detected and analyzed the reaction products, and used SYBR Gold as an indicator for 1% agarose gel electrophoresis for verification analysis. From figure 2 (A) It can be seen that when there is thymine DNA glycosylase (TDG), the characteristic band of the reaction product can be seen, and when no thymine DNA glycosylase (TDG) is added, no characteristic band appears. This is because thymine DNA glycosylase (TDG) excises mismatched thymines and initiates subsequent uracil excision-mediated circular rolling circle exponential amplification. In order to prove the dual signal amplification reaction mediated by enzyme repair, we added the signal probe and detected the reaction process with real-time fluorescence. Such as figure 2 As shown in (B), in the presence of thymine DNA glycosylase (TDG), the fluorescence intensity increased rapidly in the first 10 minute...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com