Anchor primer for efficient and rapid amplification of cDNA ends and amplification method
An anchoring and rapid technology, applied in DNA preparation, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of low amplification efficiency, low gene abundance, poor specificity, etc., and achieve good specificity and amplification. High efficiency and easy operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
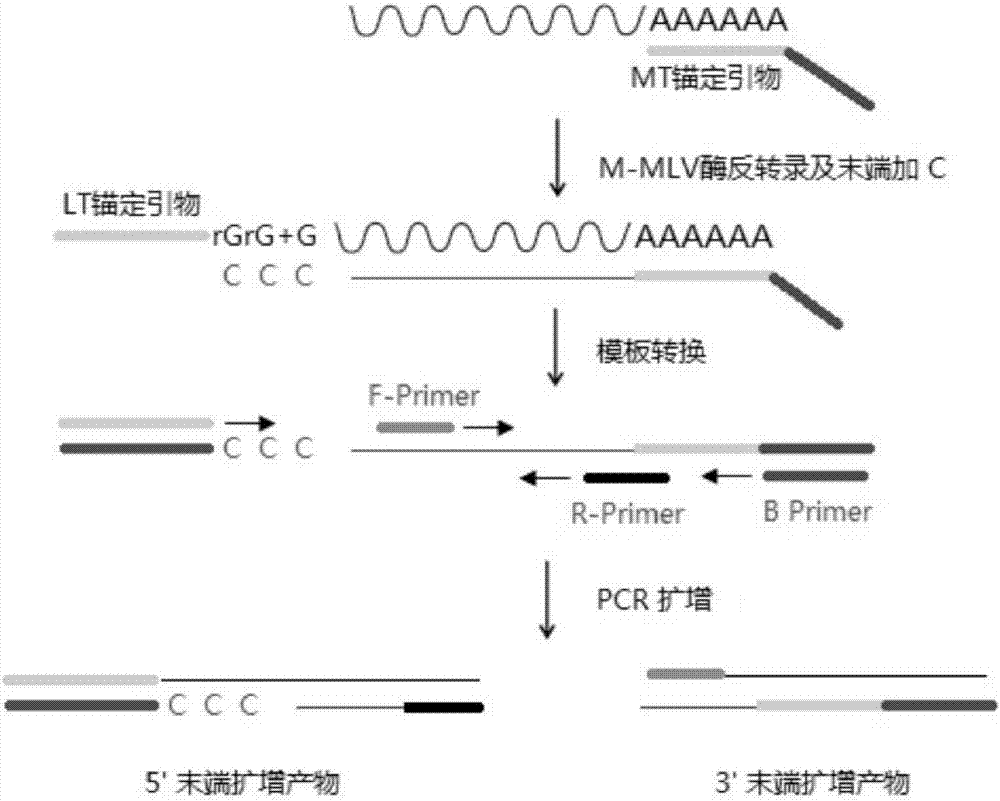
[0022] A method for efficiently and rapidly amplifying cDNA 5' and 3' ends, comprising the steps of:
[0023] a. Design a specific locking primer connected to the 5' end of the cDNA, that is, the anchor primer, denoted as LT. There are 3 guanines at the 3' end of the sequence, the first two guanines are riboguanosine, and the last guanine base is locked nucleic acid modified guanine (LNA). The primer can form 3 'Stem-loop structure protruding from the terminal portion. The special modification at the end of the adapter sequence and its own structure can significantly improve the anchoring efficiency of the locked primer;
[0024] b. Design an anchor sequence combined with the poly(A) tail at the 3' end, denoted as MT. The end of the sequence contains 30 thymines and does not form a hairpin structure by itself;
[0025] c. Design a primer that is partially complementary to the MT sequence, denoted as B;
[0026] d. Design two specific primers for the target gene, wherein th...
Embodiment 227227
[0029] Example 2 Rapid amplification of 27227 lncRNA 5' and 3' ends
[0030] 27227lncRNA is a long non-coding RNA, and the function and gene name of this long non-coding RNA are still uncertain. The accession number of its UCSC database is known as TCONS_00027227, named 27227 by itself, and its chromosome location position: chr19: 22025306-22035178.
[0031] According to the method described in Example 1, the 5' and 3' ends of 27227lncRNA were amplified, and its key parts were:
[0032] 1. Nucleic acid sequence design
[0033] The specific nucleic acid sequence used in this embodiment, the specific sequence is as follows:
[0034] The specific nucleic acid sequence used in the present embodiment of table 1
[0035]
[0036] Wherein, rG in the above table is monodeoxyguanine (riboguanosine), +G is locked nucleic acid modified guanine (LNA), and then the reverse transcription reaction system is prepared according to Table 2:
[0037] Table 2 Reverse transcription reaction s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com