Method of assistant efficient breeding of corns cooperatively using two molecular markers of SSR and SNP
A molecular marker, corn technology, applied in the field of molecular biology and genetic breeding, can solve the problems of difficult to achieve high-throughput sites and high detection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Method for using two molecular markers of SSR and SNP in combination to carry out corn assisted high-efficiency breeding
[0034] 1Materials and methods
[0035] 1.1 Test materials
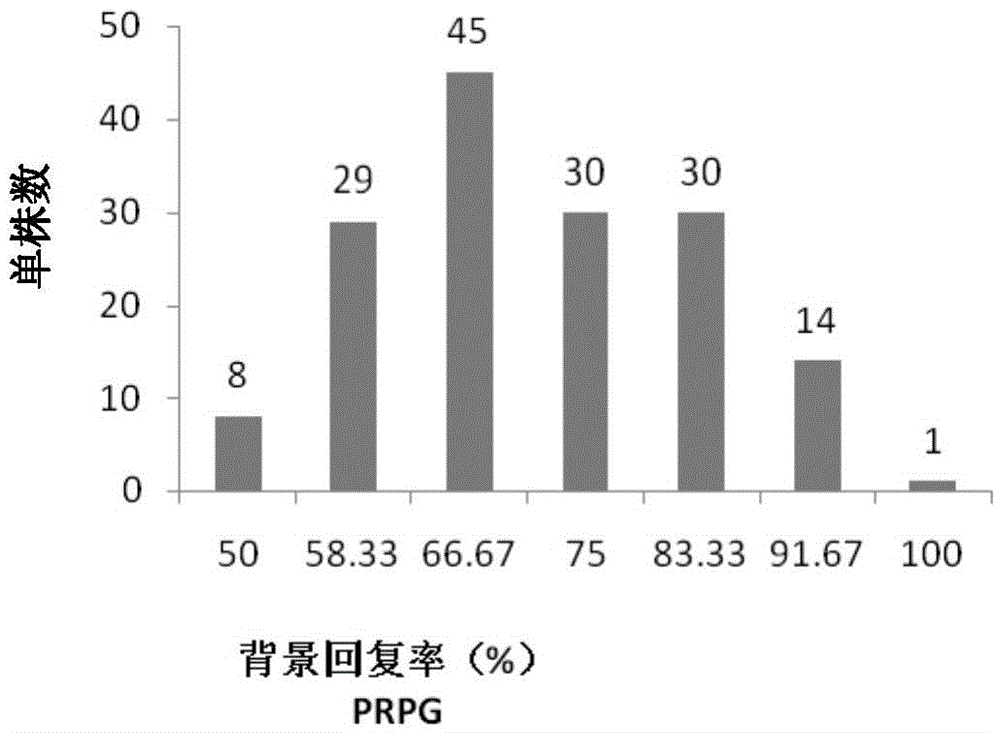
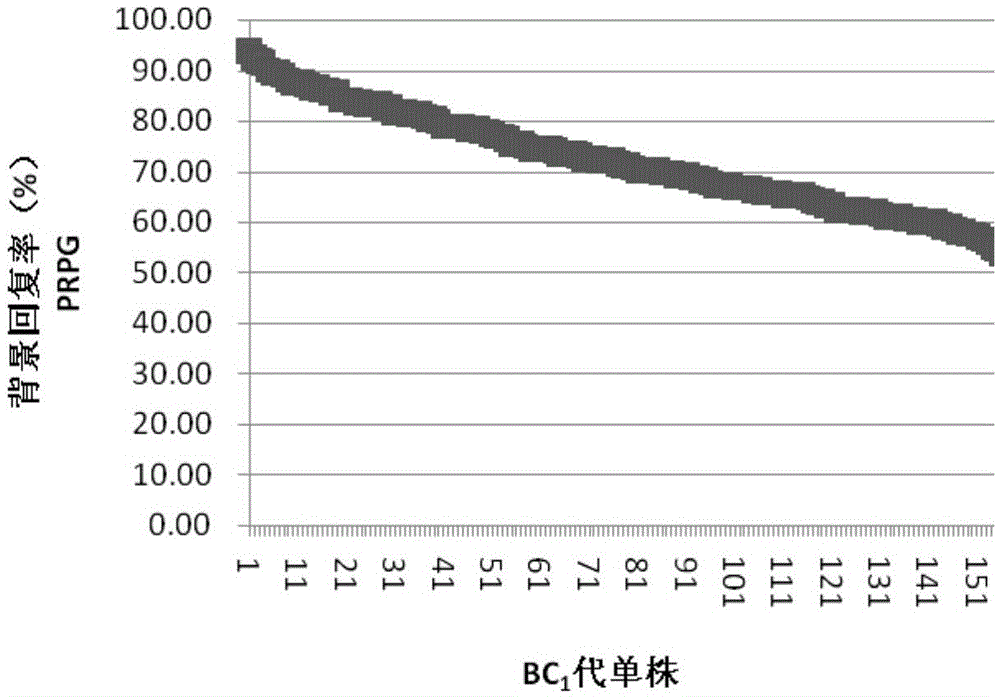
[0036] The maize inbred line Jing SD121 has high combining ability and excellent comprehensive characters, but has the disadvantages of high ear position and poor lodging resistance. With low ear position and good lodging resistance, the inbred line Jing SD127, which is closely related to Jing SD121, was used as the donor parent, and Jing SD121 was used as the recurrent parent to improve the lodging resistance of Jing SD121 through backcrossing. In this example, 157 BC strains initially selected by field phenotype selection were selected. 1 F 1 The generation backcross group is the research material.
[0037] 1.2 DNA extraction
[0038] The seedlings of Jing SD127 and Jing SD121 were grown in indoor sand culture, and DNA was extracted at the 3 to 5 leaf stage. BC 1 F 1 Generation field s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com