A kind of her2 nucleic acid aptamer and its application
A nucleic acid aptamer and nucleotide sequence technology, which can be used in pharmaceutical formulations, DNA/RNA fragments, gene therapy, etc., and can solve problems such as reducing the binding ability of nucleic acid aptamers and target molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] The preparation of embodiment 1HB3-1, HB3
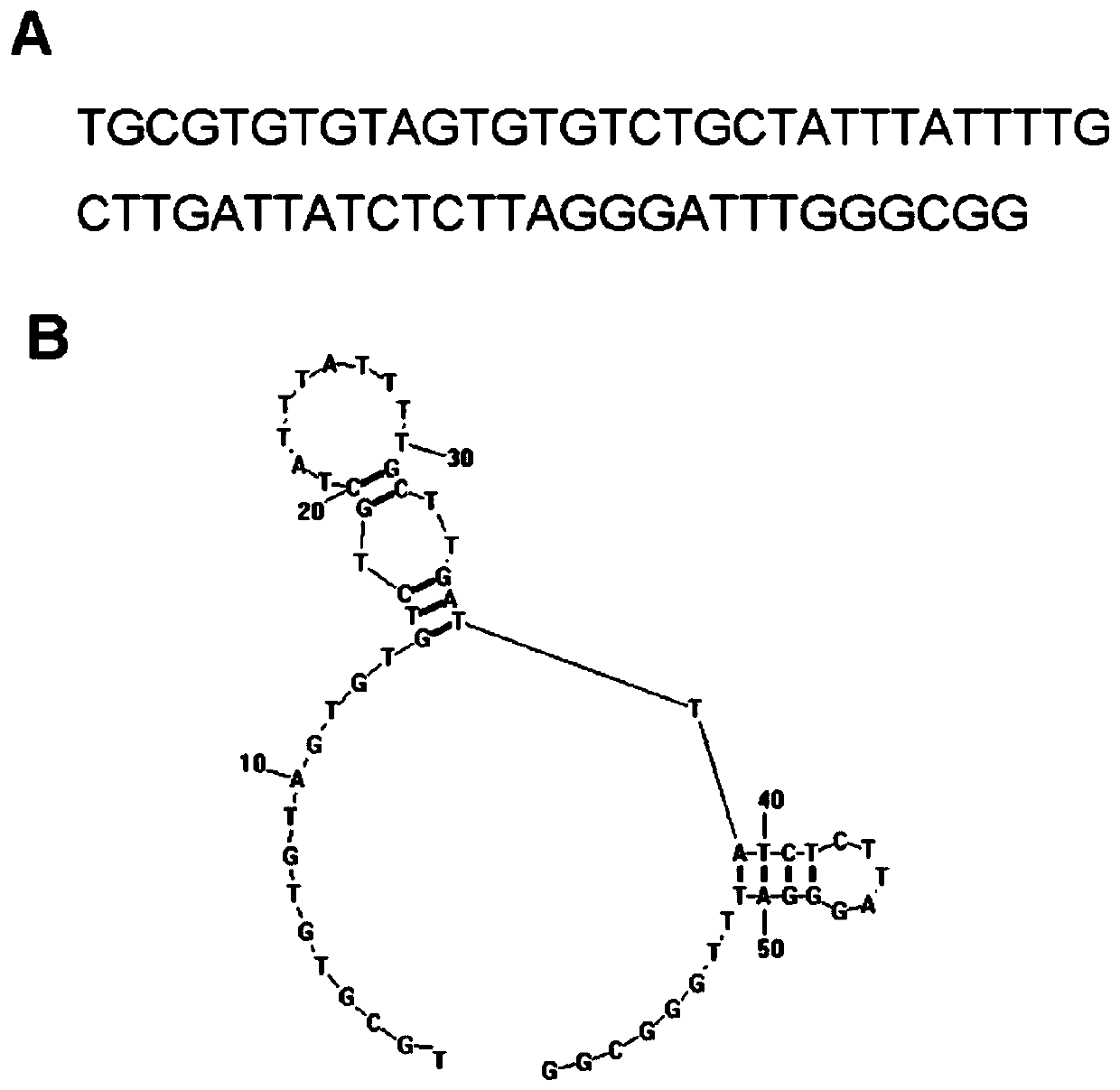
[0059] In the present invention, the core sequence of the HER2 nucleic acid aptamer is a single-stranded nucleotide with a length of 43 bases, and its nucleotide sequence is: 5'GTGTGTCTGCTATTTATTTTGCTTGATTATCTCTTAGGGATTT3'; the preferred HER2 nucleic acid aptamer is 59 bases The length of single-stranded nucleotides, the sequence is: 5'TGCCCGTGTCCCGAGGAGTGCCCTATTTTGCTTGATTATCTCTAAGGGATTTGGGCGG-3'; more preferably, when synthesizing, all the bases A in the above-mentioned sequence are modified with sulfur to obtain a sulfur-substituted HER Nucleic acid aptamers, named respectively (the core sequence is thiolated (HB3-1), the preferred sequence is thiolated (HB3)), the above sequences were synthesized by Invitrogen Company. The structure of HB3 aptamer is as figure 1 In the figure, A is the primary sequence and B is the secondary structure.
[0060] Hereinafter, if there is no special statement, the terms "HB3" and "thio-HB3" ...
Embodiment 2
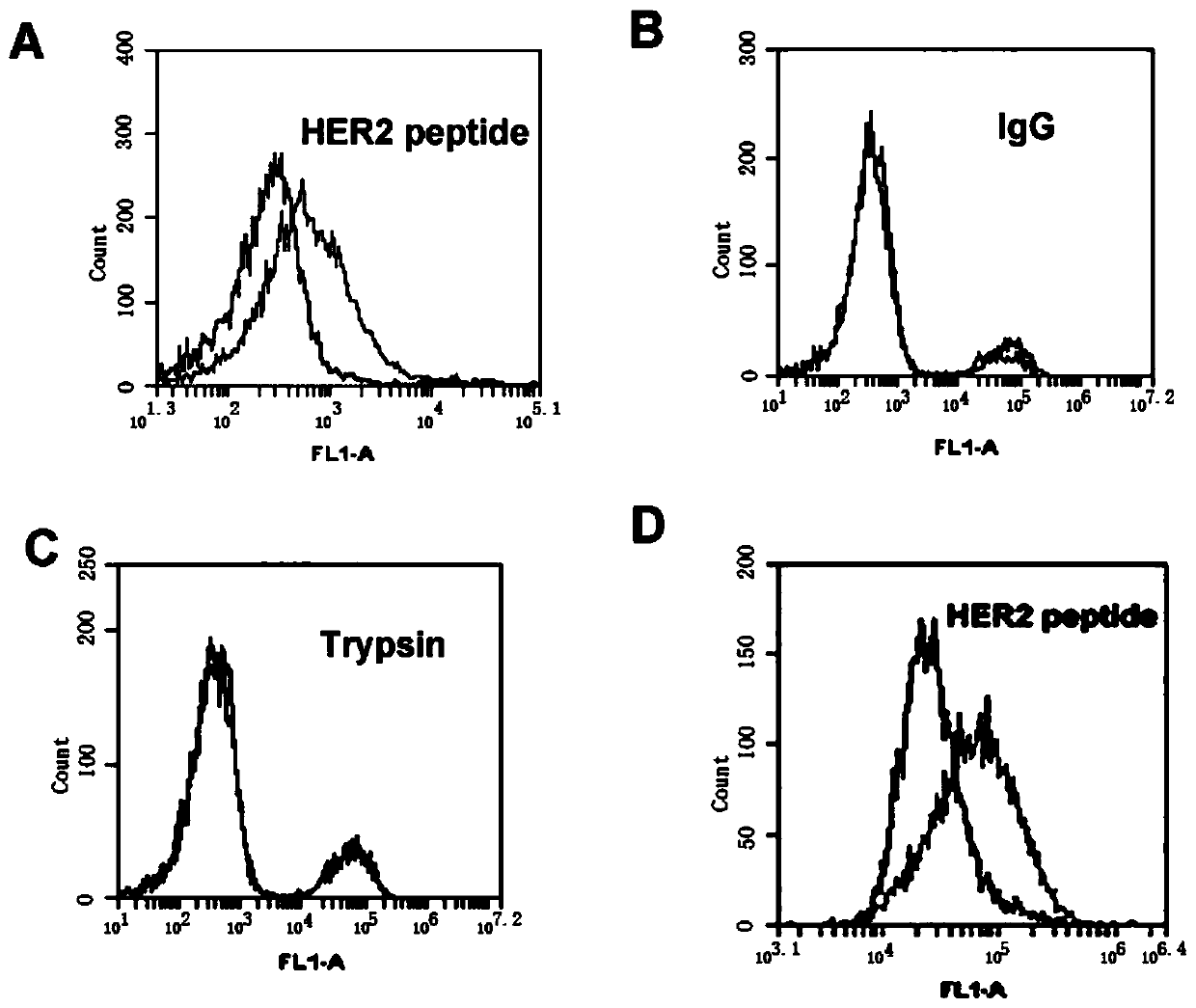
[0061]Example 2 The ability of HB3-1 to bind to HER2 polypeptide
[0062] HB3-1 modified with biotin (biotin) at the 3' end and fluorescein (FITC) at the 5' end was mixed with HER2 polypeptide, trypsin and HER2 polypeptide in 200 μl of binding buffer (PBS buffer with pH 7.4), respectively. The IgG-coated magnetic beads were reacted at 37°C for 30 min, washed twice with PBS, and analyzed by flow cytometry. The primary structure of the modified HB3-1 is as follows:
[0063] 5'FITC-GTGTGTCTGCTATTTATTTTGCTTGATTATCTCTTAGGGATTT-biotin-3'. Base A in the sequence is a sulfur modification.
[0064] Under the same conditions, we detected the binding of base A to HER2 polypeptide when it was not thiomodified.
[0065] The result is as figure 2 , where (A) the binding of HER2 polypeptide-coated magnetic beads to FITC-labeled HB3-1; (B) the binding of IgG antibody-coated magnetic beads to FITC-labeled HB3-1; (C) trypsin-coated beads Binding of magnetic beads to FITC-labeled HB3-1; (D...
Embodiment 3
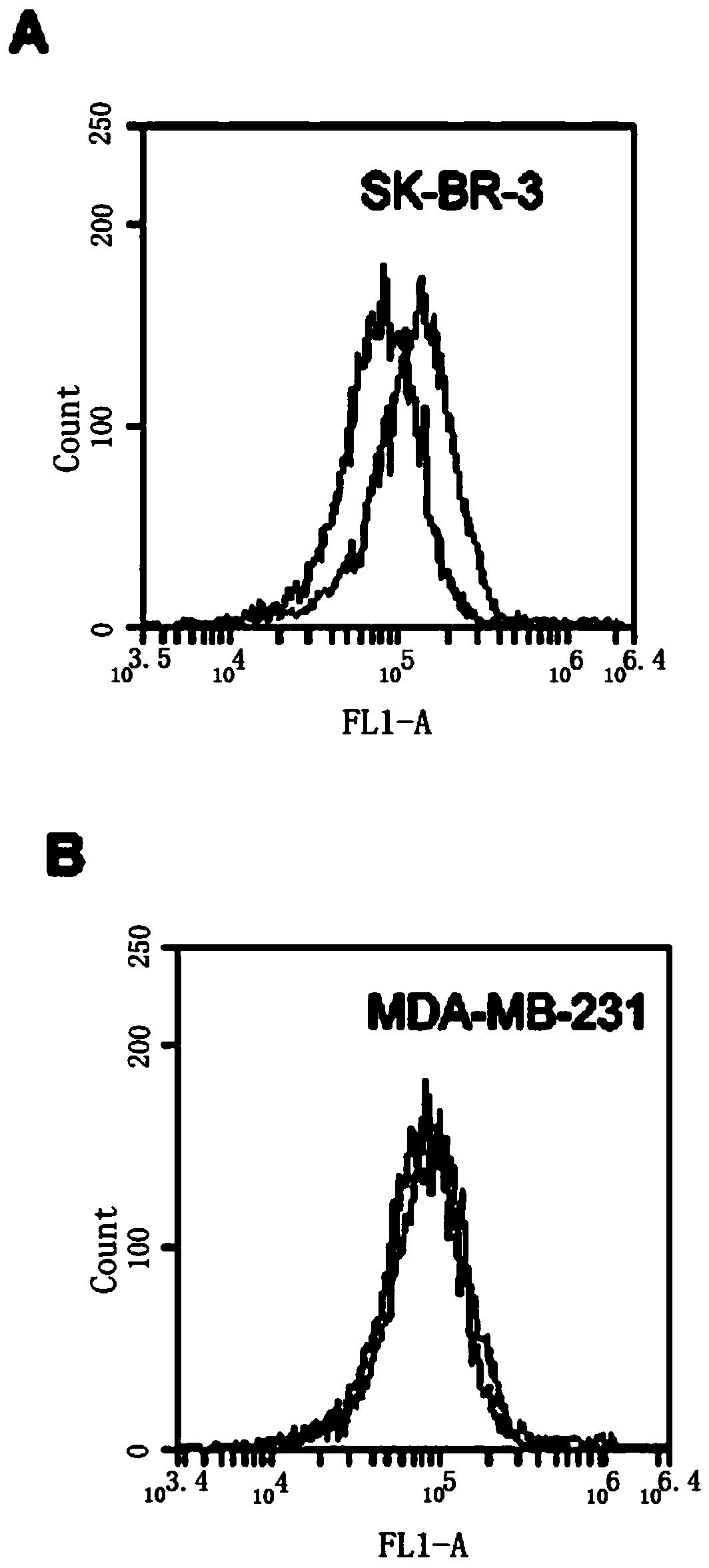
[0066] Example 3 The ability of HB3-1 to bind to HER2-positive tumor cells
[0067] SK-BR-3 cells were cultured in modified RPMI1640 containing 15% FBS, MDA-MB-231 cells were cultured in DMEM high glucose medium containing 10% FBS, 100 U / ml penicillin and 100 mg / ml streptomycin, All cells were kept at 37°C, 5% CO 2 The cells were cultured in an incubator, and the cells used in the following experiments were all cells in the logarithmic growth phase.
[0068] Scrape 5×10 separately 5 The SK-BR-3 and MDA-MB-231 cells were reacted with HB3-1 (biotin modified at the 3' end and fluorescein at the 5' end) in 200 μl binding buffer (PBS) at 37 °C for 30 min, Washed twice with PBS and analyzed by flow cytometry. see the results image 3 , (A) HER2 positive SK-BR-3 cells. (B) HER2 negative MDA-MB-231 cells. The black curve is the control fluorescence signal generated by the random library single-stranded DNA, and the gray curve is the fluorescence signal generated by thio-HB3-1. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com