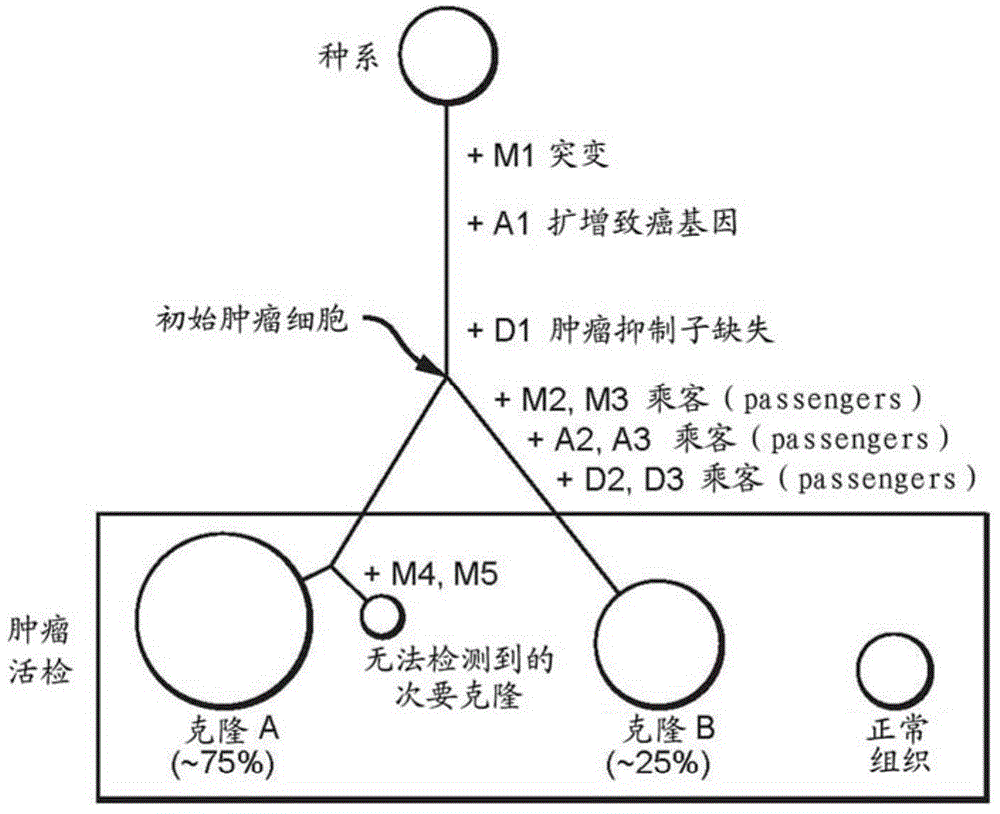
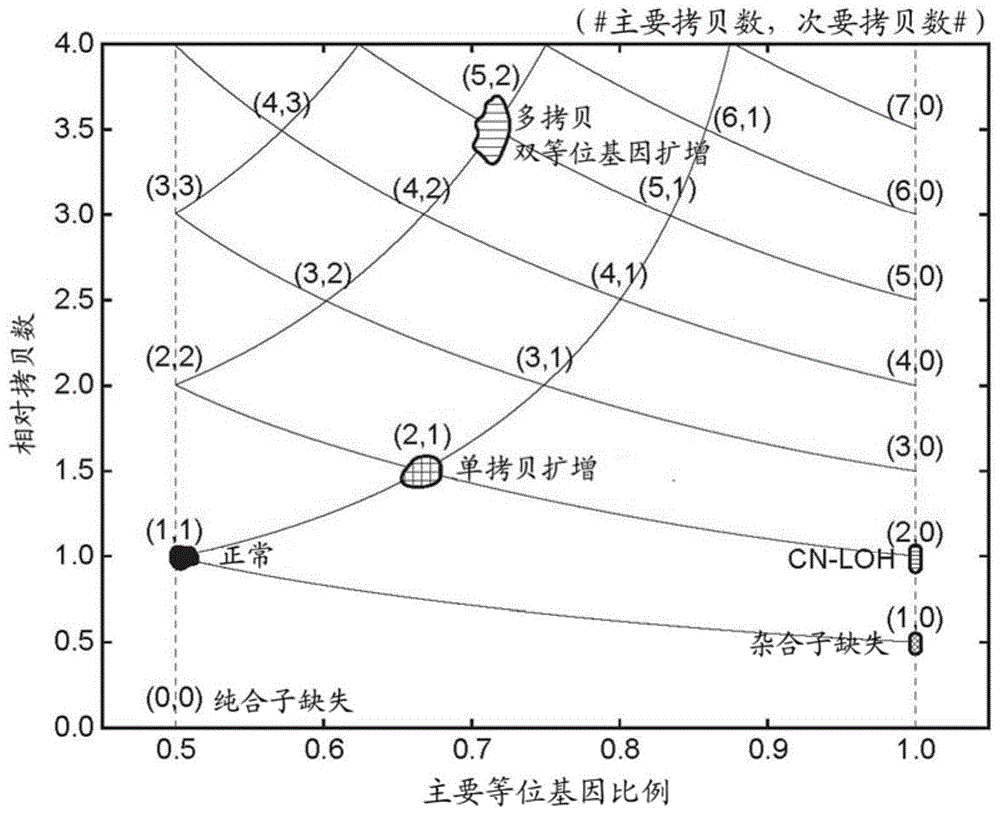
Systems and methods for tumor clonality analysis
A clonality and tumor technology, applied in the field of computational analysis of genomic data, can solve the problems of inability to identify tumors, inability to identify the relationship between clonality and cell population clones, inability to gain insight into clonality and potential therapeutic efficacy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0103] GBM (glioblastoma): Twelve genome-wide GBM samples were processed as described above for the determination of normal contamination levels and clonality present in each tumor biopsy. For the other 5 genome-wide GBM samples discussed in the previous section, the relative coverage and allele ratios generated by BamBam would have too much variability to be analyzed by these methods. Table 1 summarizes the results of the clonality analysis.
[0104]
[0105] Surprisingly, only 3 GBM tumor samples were found to be monoclonal, while the other 9 samples included at least two predominant clones. For 7 GBM tumors, the exact mixture of clones was determined, while the remaining 5 tumors were visually inspected to determine their clonality.
[0106] The results of two clones GBM-06-0145 and GBM-06-0185 are as follows Figure 7A with 7B shown. Relative coverage and allele ratio data for the two samples transformed by the best fit parameters described above showed a close fit ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com