A method for detecting chromosomal microdeletions and microduplications in human embryos
A micro-deletion and micro-repetition technology for chromosomes, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of low throughput, low resolution, long DNA fragments, etc., and achieve low cost and high sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
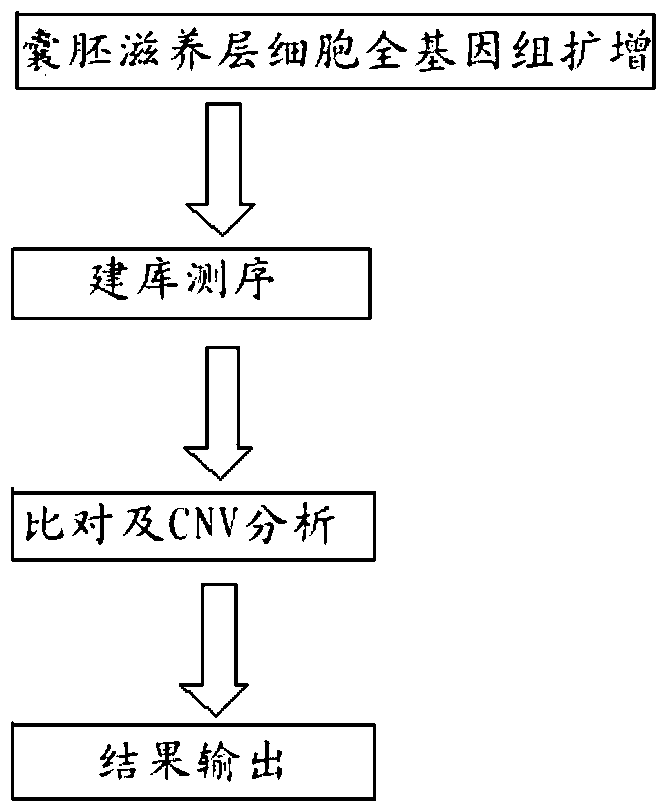
Image
Examples
Embodiment 1
[0049] 1. Material:
[0050] 1. Specimen: The blastocyst stage cells biopsied from a cooperative hospital. The source of the sample was two embryo samples of IVF. The parental karyotype was checked as a carrier of balanced translocation, and he was pregnant with an abnormal fetus and caused a miscarriage.
[0051] 2. Reagents: Qiagen's whole genome amplification kit, MDA product detection kit, MDA product purification kit, life library construction kit, Agencourt®AMPure®XP magnetic beads, life template preparation kit, life sequencing kit.
[0052] 3. Instruments: PCR instrument, agarose gel electrophoresis system, ion torrent's PGM sequencing platform.
[0053] 4. Consumables: 1.5ml, 0.2ml imported centrifuge tube, Raining pipette and tip with filter element.
[0054] 2. Operation steps:
[0055] 1. Trophoblast cell biopsy
[0056] Using the two-step method, the first step is to perforate the zona pellucida on all embryos at D3 before the biopsy. On D5 or D6, perform trophoblast cell col...
Embodiment 2
[0153] 1. Material:
[0154] 1. Specimen: blastocyst stage cells biopsy from a cooperative hospital. The source of the sample was three embryo samples of IVF. The parent had repeated abortions before.
[0155] 2. Reagents: Qiagen's whole genome amplification kit, MDA product detection kit, MDA product purification kit, life library construction kit, Agencourt®AMPure®XP magnetic beads, life template preparation kit, life sequencing kit.
[0156] 3. Instruments: PCR instrument, agarose gel electrophoresis system, ion torrent's PGM sequencing platform.
[0157] 4. Consumables: 1.5ml, 0.2ml imported centrifuge tube, Raining pipette and tip with filter element.
[0158] 2. Operation steps:
[0159] 1. Trophoblast cell biopsy
[0160] Using the two-step method, the first step is to perforate the zona pellucida on all embryos at D3 before the biopsy. On D5 or D6, perform trophoblast cell collection on the two high-quality blastocysts formed under the microscope. Collect 5-10 trophoblast cells a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com