Heterozygous genome processing method
A processing method and genome technology, applied in the fields of genome bioinformatics analysis and genetic engineering, can solve the problems of long cycle, high cost, restricting species genome interpretation, etc., and achieve the effect of shortening cycle and reducing cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0060] The objects, features and characteristics of the present invention, as well as the operation method of the relevant elements of the structure and the function and combination of parts will become more apparent when reading the following description and the above claims with reference to the accompanying drawings. It is to be expressly understood that the drawings are for purposes of illustration and description only and are not intended as limitations of the invention.
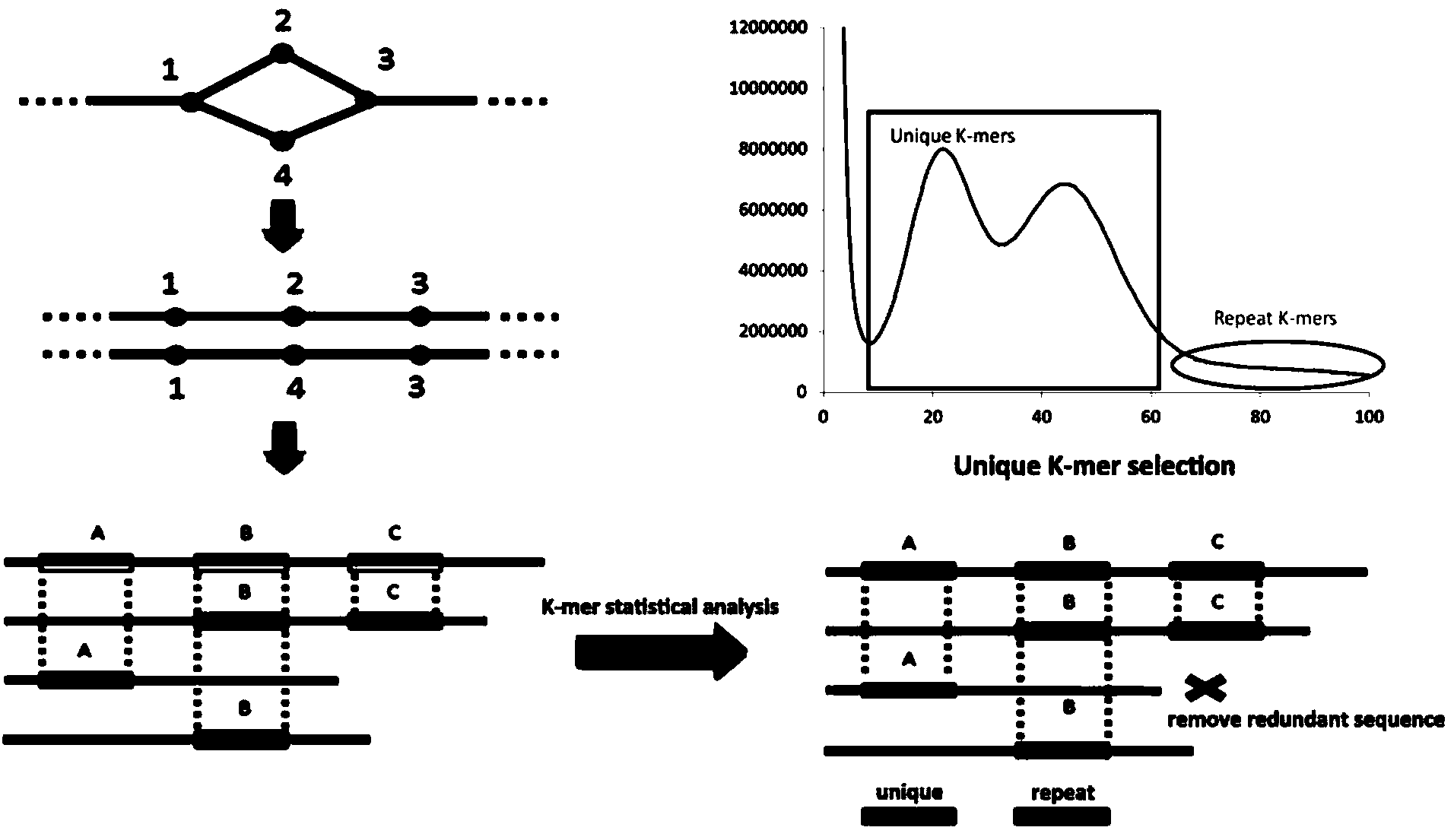
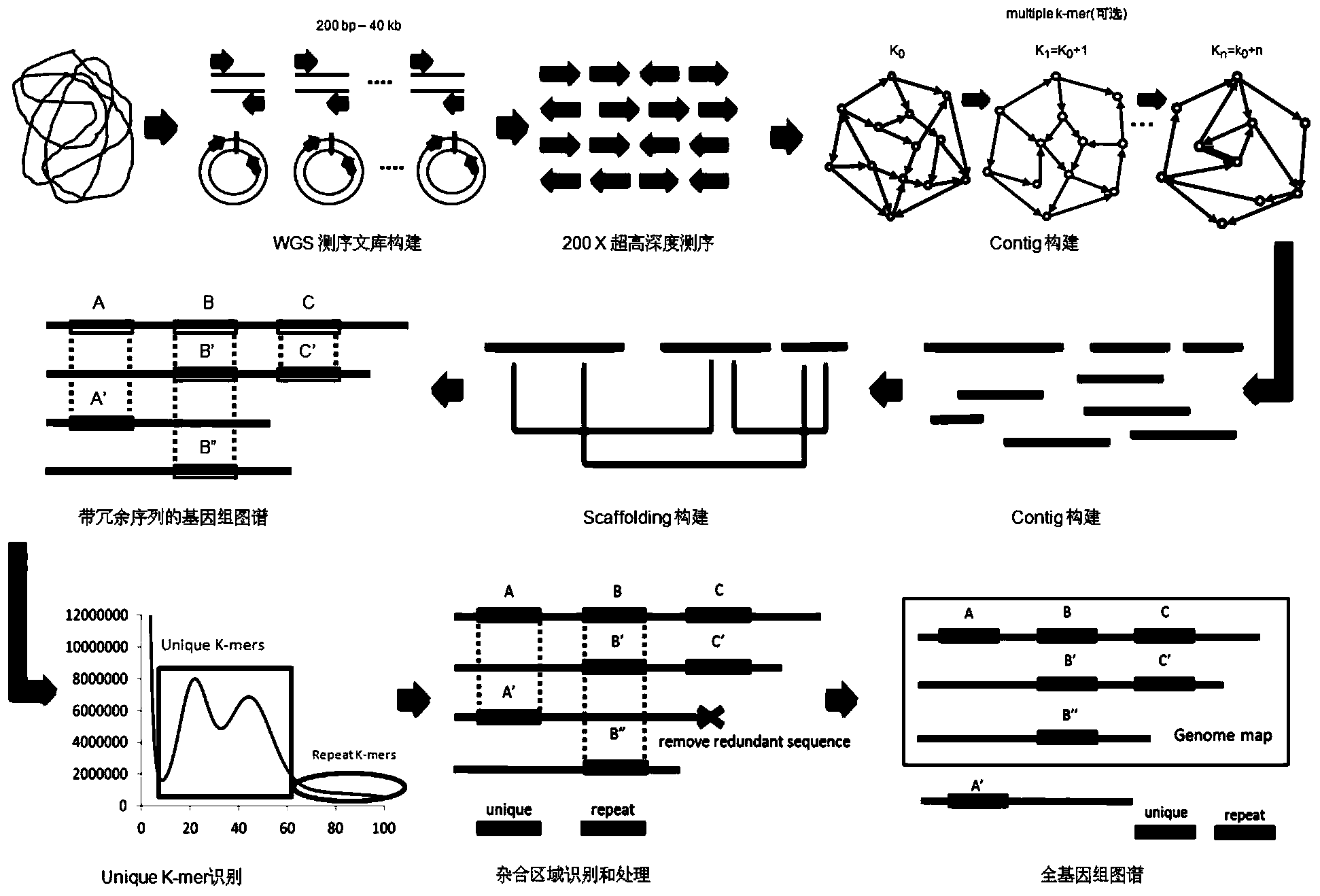
[0061] In the heterozygous genome processing method of the present invention, without any reference sequence, based on the next-generation high-throughput sequencing technology, the target species is sequenced at an ultra-high depth (generally more than 200 times), and then the new version of SOAPdenovo2 assembly software is used , combined with heterozygous recognition processing, to obtain the whole genome sequence map of the target species. Embodiments of the present invention may include whole genom...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com