A multiplex PCR detection kit and method for rapidly identifying pathogenic Pseudomonas ayumi
A sweetfish Pseudomonas, detection kit technology, applied in the directions of microorganism-based methods, microorganism determination/inspection, biochemical equipment and methods, etc. The environmental isolates, lack of specificity and other problems, to achieve the effect of short detection time, simple instrument requirements, labor and financial savings
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] A multiplex PCR detection kit for pathogenic Pseudomonas ayumi, the kit specifically consists of the following reagents:
[0035] TaqDNA polymerase 0.05mL, 10×PCR buffer 0.5mL, deoxyribonucleoside triphosphate mixture 0.25mL, redistilled water 5mL, 3 pairs of specific primers with a concentration of 10μM respectively 200μL, Pseudomonas RNA polymerase The upstream and downstream primers P1 and P2 of the subunit gyrB, the upstream and downstream primers P3 and P4 of Pseudomonas ayumi gyrB, and the upstream and downstream primers P5 and P6 of the pathogenic Pseudomonas ayumanus type III secretion system regulator exsA.
[0036]Among them, relatively degenerate primers were designed based on the internal conserved sequences of the gyrB genes of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas ayumi with gene accession numbers CP000094.2, KC189956.1, and AB178859.1 , the target sequence length is 745bp; among them, the upstream and downstream primers of Pseudomona...
Embodiment 3
[0053] According to the method of Example 2, the DNA template solution extracted in the method was serially diluted in multiples, with a concentration ranging from 1050 ng / μL to 1 ng / μL, and the same multiplex PCR detection as in Example 2 was performed, and it was clear within the concentration range of ≥4 ng / μL 3 specific purpose bands were amplified accurately, showing that the sensitivity of DNA that can be detected by the multiplex PCR established by the present invention is 4ng / μL, and the results are as follows: figure 2 As shown, where M is the DNA molecular weight standard DL2000, 1-10 are template DNA concentrations of 1050ng / μL, 525ng / μL, 258ng / μL, 129ng / μL, 65ng / μL, 32ng / μL, 16ng / μL, 8ng / μL, respectively. Multiplex PCR detection results at μL, 4ng / μL, 2ng / μL, 1ng / μL, lane 11 is negative control.
Embodiment 4
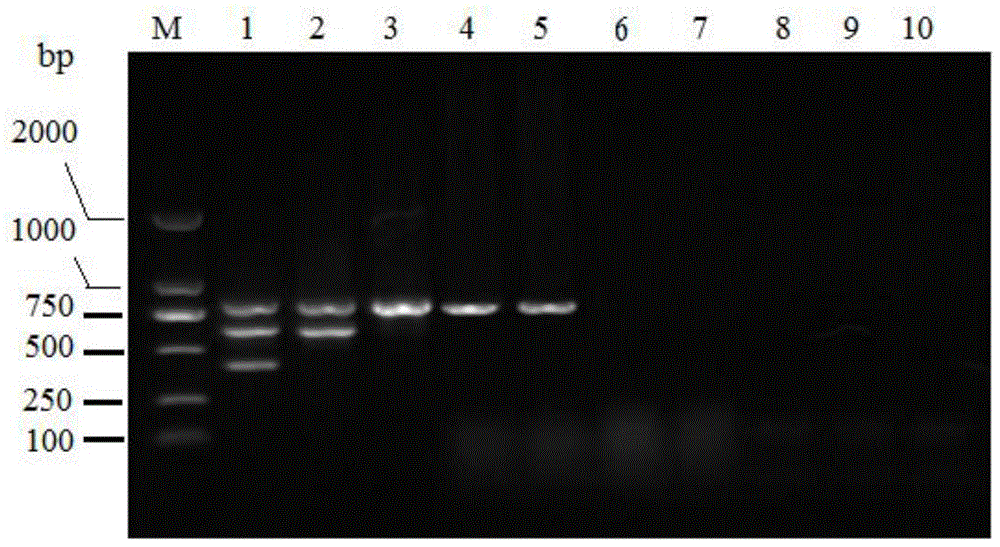
[0055] Respectively for Pseudomonas aeruginosa, Pseudomonas fluorescens, Pseudomonas putida, Pseudomonas ayumi (pathogenic strains and environmental isolates, the environmental isolates are provided by the Marine Microorganism Collection Center of the State Oceanic Administration), Vibrio harveyi, Vibrio parahaemolyticus, Vibrio alginolyticus and Aeromonas hydrophila etc. were extracted template DNA according to the method in Example 2, and carried out the same multiple PCR detection, and the electrophoresis results were as follows: image 3 , indicating that the multiplex PCR established by the present invention can specifically detect Pseudomonas ayumi, and at the same time distinguish pathogenic strains from environmental isolates. Among them, M is the DNA molecular weight standard DL2000, and lanes 1-9 represent Pseudomonas ayuyui NB2011, environmental isolates of Pseudomonas ayumi (MCCC1A00022), Pseudomonas putida (CGMCC1.8092), Pseudomonas fluorescens bacterium (CGMCC1.6...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com