Gradient intensity promoter of gluconobacter oxydans
A technique for oxidizing glucose, a strong promoter, used in the field of genetic and metabolic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
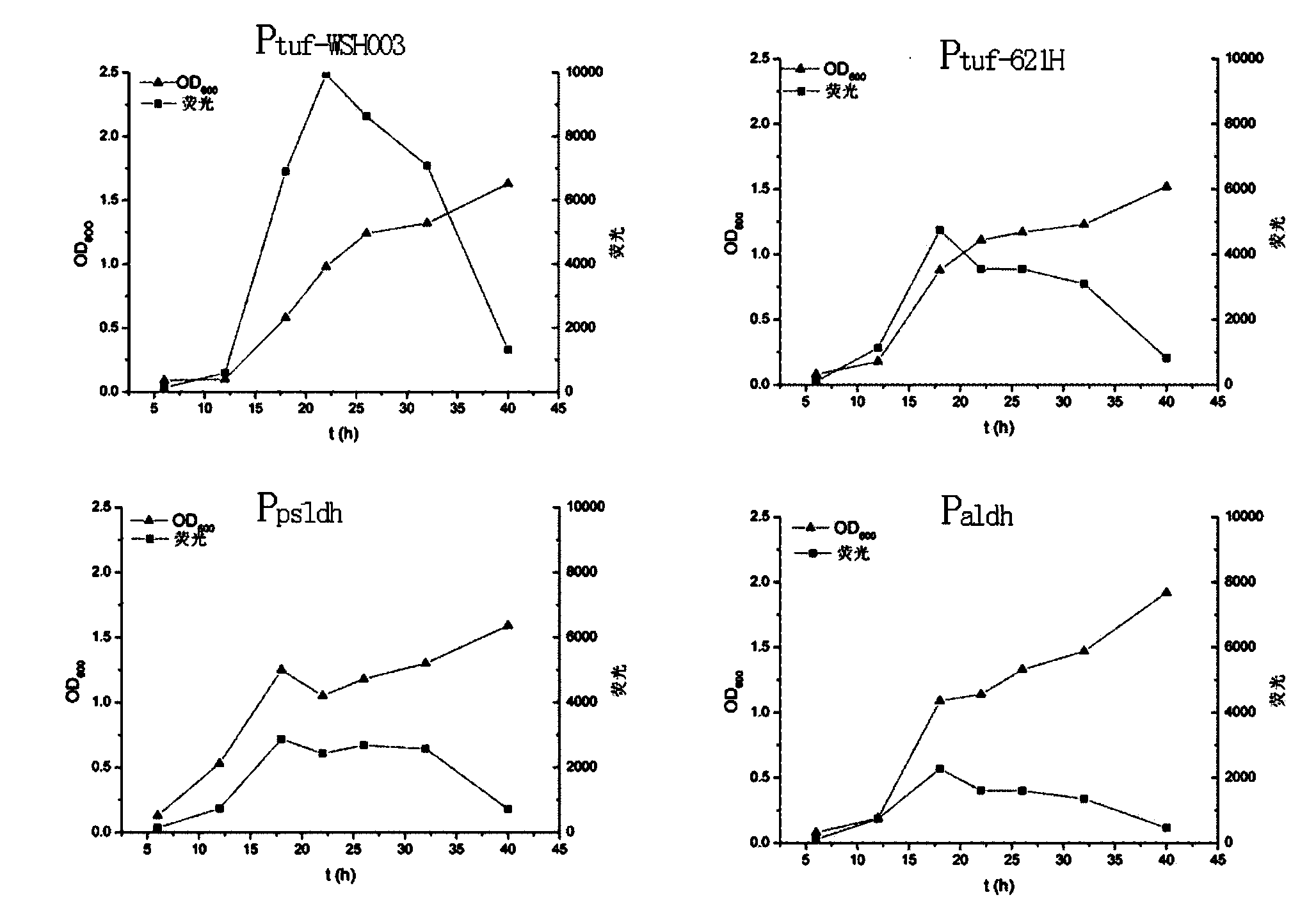
[0022] Example 1 Selection of promoters
[0023] G oxydans contain a variety of important dehydrogenases related to the metabolism of glucose, glycerol, sorbitol, etc. These enzymes may have potential strong constitutive expression promoters, such as sorbitol dehydrogenase (sorbitol dehydrogenase), membrane-bound aldehyde Promoters of dehydrogenase (aldehyde dehydrogenase), membrane-bound glucose dehydrogenase (membrane-bound glucose dehydrogenase) and the like. In addition, the P in the strongest Gluconobacter oxydans 621H reported in the literature was selected tuf-621 H served as a control. According to the G.oxydans WSH-003 genome sequence information (GenBank accession NO.AHKI00000000), select all nucleotide fragments between the target gene and the ORF (open reading frame) of the previous gene, and then according to the promoter software ( http: / / www.softberry.com ) to evaluate the score of the promoter region, select a promoter with a higher score, and use EGFP as a ...
Embodiment 2
[0024] Example 2 Construction of expression vector pBBR1MCS2-promter-egfp
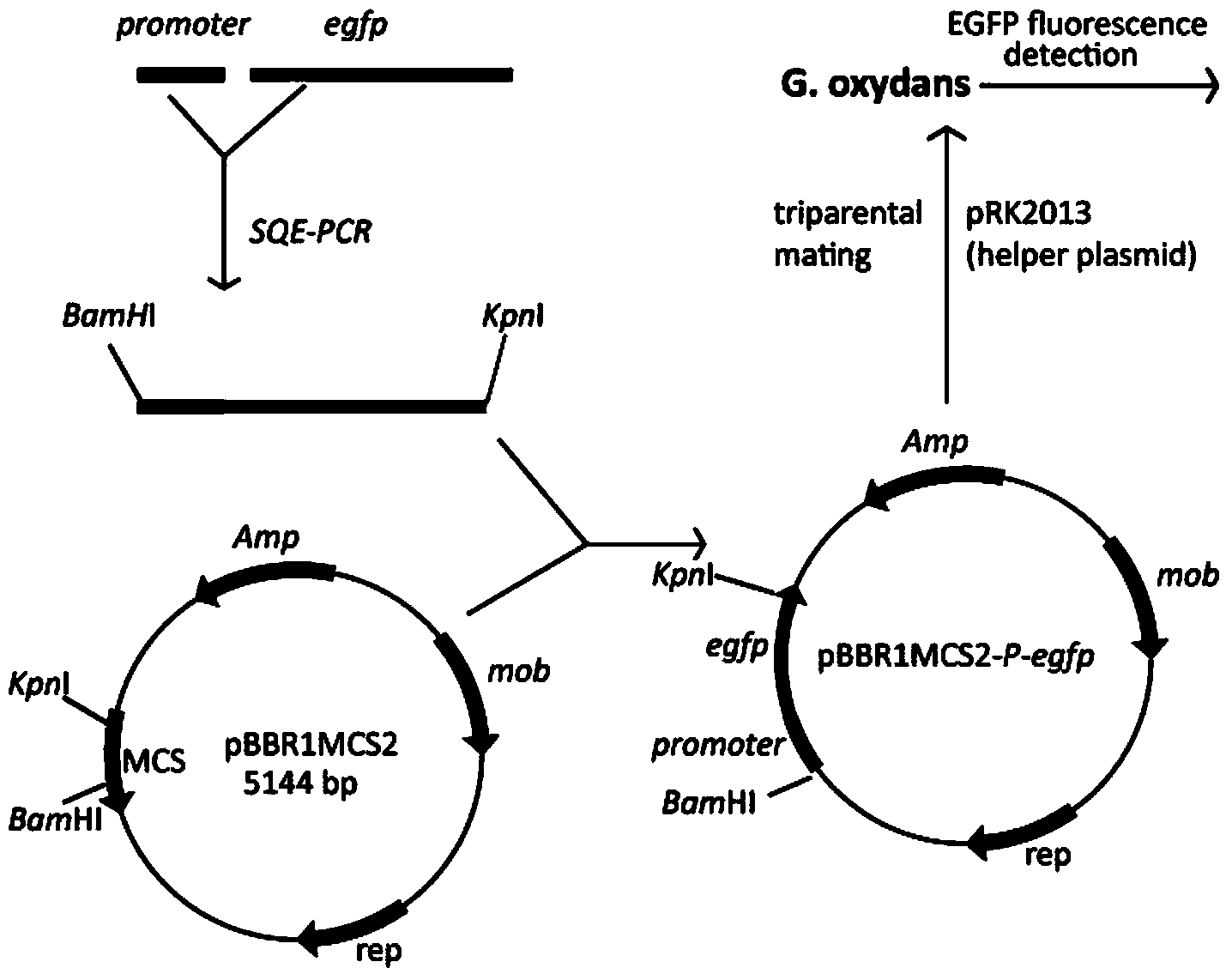
[0025] According to the genome sequence information of G.oxydans WSH-003 (GenBank accession NO.AHKI00000000), fusion PCR primers were designed to amplify the highly scored promoter nucleotide sequence and the gene sequence of enhanced green fluorescent protein egfp (egfp gene sequence includes Terminator, the sequence is shown in SEQ ID NO.7), and then connected by fusion PCR to obtain promoter-egfp with restriction sites at both ends, perform T-A cloning and sequencing, and connect the positive clones with the same enzyme The broad-hosted vector pBBR1MCS2 with cutting sites was constructed into the expression vector pBBR1MCS2-promter-egfp( figure 1 ).
Embodiment 3
[0026] Example 3 Construction of Fluorescent Protein Recombination Detection Strain
[0027] The cloned expression vector pBBR1MCS2-promoter-egfp verified by sequencing was introduced into G. oxydans WSH-003 (preserved in the Industrial Microbiology Resource and Information Center of Chinese Universities, Jiangnan University, No. CICIM-CU B7004) by the method of three-parent hybridization. E.coli JM109 containing the recombinant expression vector was used as the donor bacterium, and E.coli HB101 containing the conjugative helper plasmid pRK2013 was used as the auxiliary bacterium. G.oxydans WSH-003 grows to OD 600 =0.9, E.coli grown to OD 600 = 0.8. Take 1ml of E.coli JM109 and E.coli HB101, mix them, centrifuge, mix and centrifuge with 2ml LB resuspended bacteria and 4ml G.oxydans WSH-003, and use 0.8ml Y-S medium (yeast extract 20g / L, Sorbet Alcohol 80g / L) to resuspend the bacteria, spread the bacteria solution on a Y-S solid medium plate containing filter paper but not c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com