Efficient method for synthesizing cDNA from RNA through reverse transcription
A reverse transcription and high-efficiency technology, applied in the direction of fermentation, can solve problems such as complex reactions, and achieve the effects of increasing the amount of amplification, increasing the amount of synthesis, and simplifying the reverse transcription reaction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
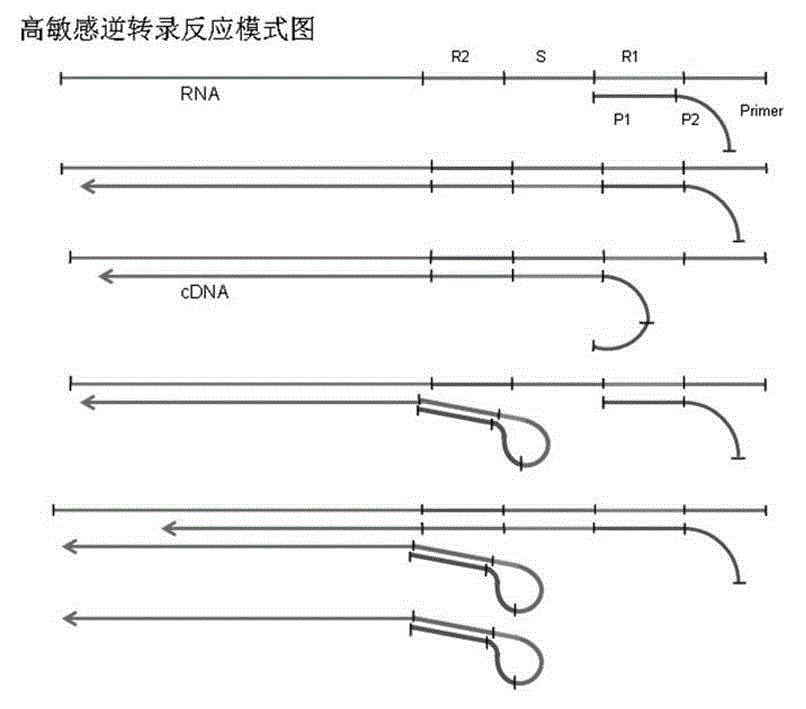
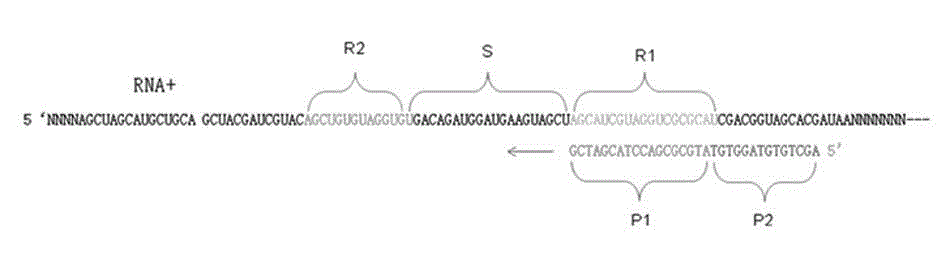
[0031] Such as figure 2 As shown, HCV viral RNA is used as the target template RNA for cDNA synthesis, and the reverse transcription compound sequence primer is: 5' ACTGCCTGATAGGG TGCACGGTCTACGAGACCTC, the 3' end of the specific primer has a specific deoxynucleotide sequence (P1) complementary to the template RNA, 19bp, which can be combined with the R1 region of the template RNA. The 5' end of the specific primer has a specific deoxynucleotide sequence (P2) that can be identical to the R2 region of the template RNA sequence, 14bp. exist figure 2 It shows that the R1 and R2 intervals of the RNA template are separated by the S region, and the length of the S region is the length of the P1 sequence plus 1 bp, that is, 20 bp.
[0032] The conventional reverse transcription primer 5' TGCACGGTCTACGAGACCTC was used as a contrast reaction.
[0033] Reverse transcription reaction setup:
[0034] The reverse transcription reaction mix of the reaction configuration includes 1 μg ...
Embodiment 2
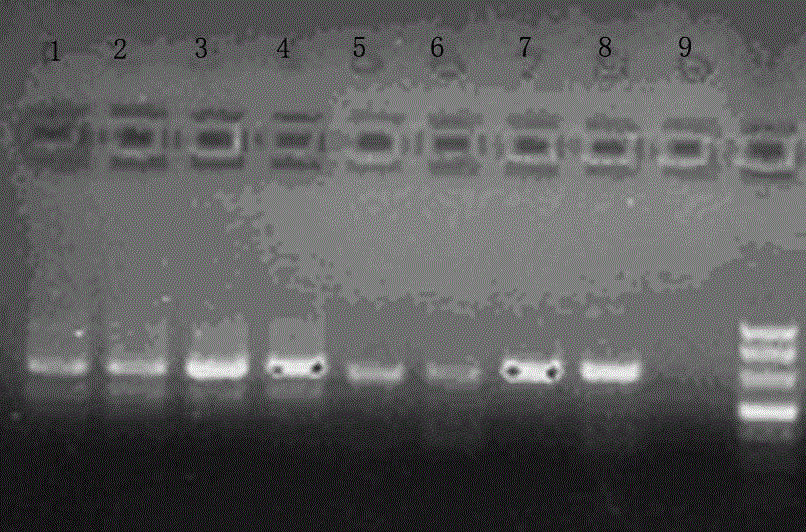
[0053] HAV RNA is used as the target template RNA for cDNA synthesis, and the specific reverse transcription compound primers and conventional reverse transcription primers described in the present invention are firstly used to carry out reverse transcription reaction to synthesize target cDNA. Real-time fluorescent PCR technology was used for PCR amplification, and the reverse transcription effects of the two primers were compared. Such as Figure 4 As shown, the reverse transcription compound sequence primer is: RT-1 5' aggaggtttggattctCCatttcaagagtccacaca, the 3' end of the specific primer has a specific deoxynucleotide sequence (P1) complementary to the template RNA, 20bp, which can be combined with the template RNA R1 area. The 5' end of the specific primer has a specific deoxynucleotide sequence (P2) that can be identical to the R2 region of the template RNA sequence, 16bp. exist Figure 4 The figure shows that the R1 and R2 regions of the RNA template are separated b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com