Method for building TALE (transcription activator-like effector) repeated sequences
A repeat sequence and tandem repeat sequence technology, applied in the field of molecular biology, can solve the problems of long exploration and adjustment, limiting the rapid and routine construction of TALE, and increasing the difficulty of experimental operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1 constructs a single side unit carrier
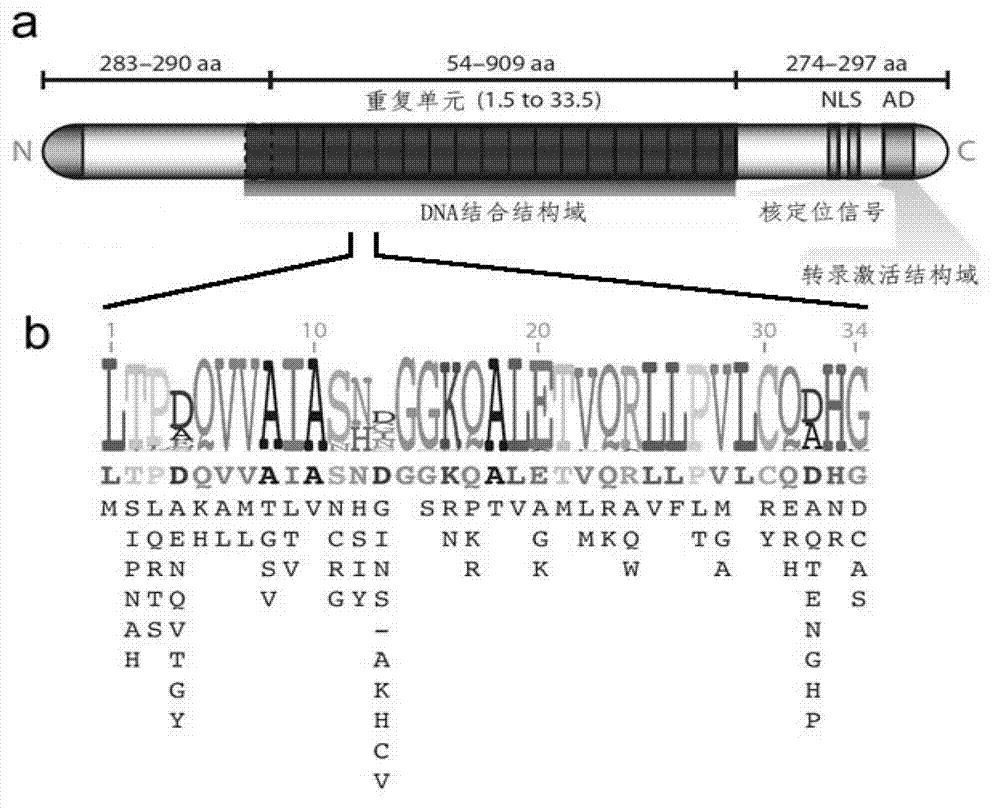
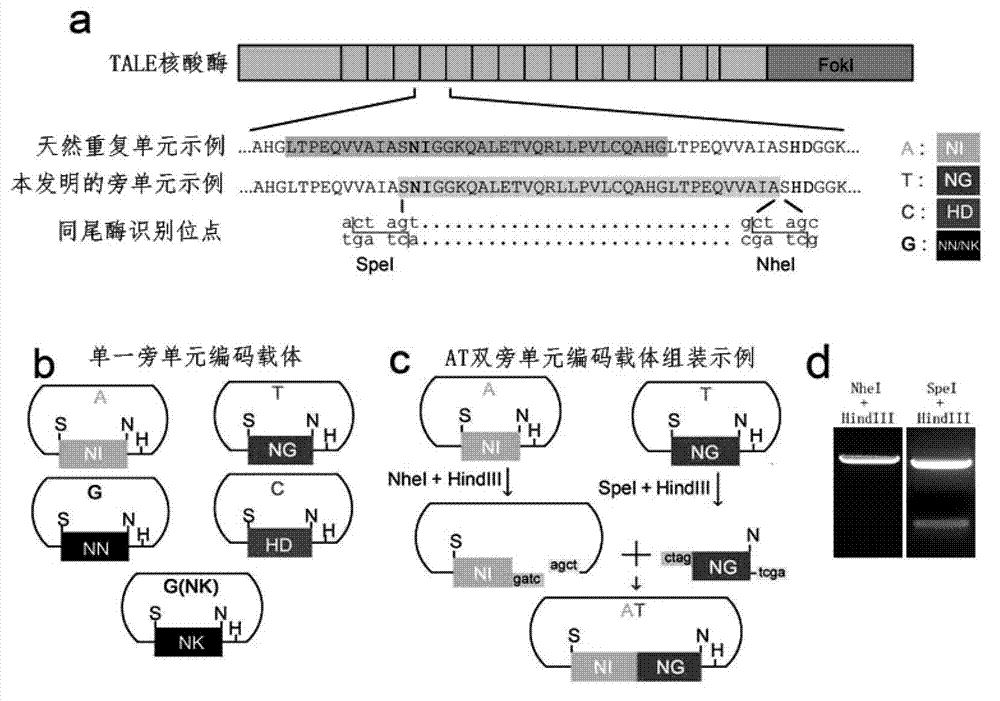
[0046] First, artificially synthesized DNA fragments (sequences listed in Table 1) encoding five kinds (classified according to the type of RVD) of the side unit sequences selected by the present invention based on the isotailase site and the typical TALE repeat unit coding sequence. The codons used by each side unit need to be carefully selected in advance to reduce the DNA sequence similarity between each side unit as much as possible. Simultaneously, since there are at least 3 possibilities, such as A, D, and E, for the amino acid residue at the +4 position in the natural repeating unit, the 15 sequence variants listed in this example are derived from five side units. Next, the five DNA fragments (15 variants) were amplified by PCR. The upstream primers are: Afwd:5'-ACTAGTAATATTGGTGGCAAACAGGCTCTTG-3'(SEQ ID No.39), Tfwd:5'-ACTAGTAATGGGGGTGGCAAACAGGCTCTTG-3'(SEQ ID No.40), Cfwd:5'-ACTAGTCATGACGGTGGCAAACAGGC TCTTG-3...
Embodiment 2
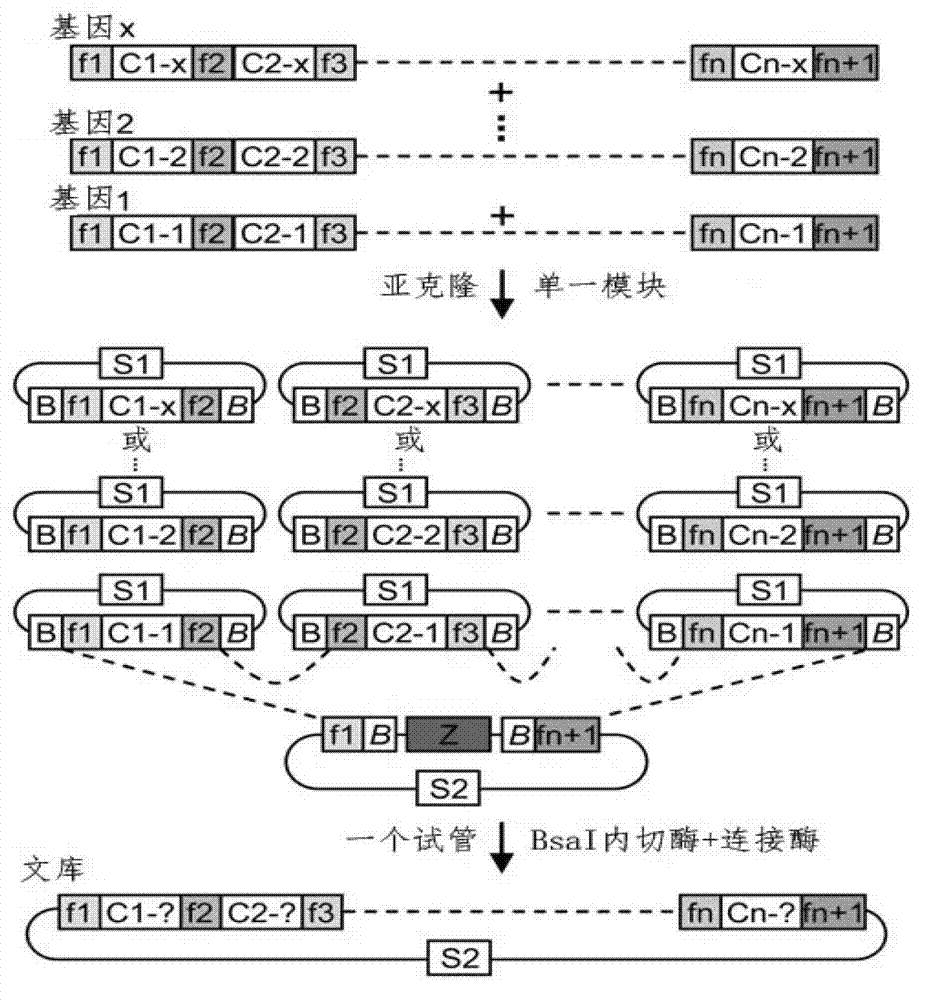
[0051] Example 2 Construction of a double side unit vector and a side unit tandem repeat vector containing n repeats
[0052] In order to construct a double side unit carrier, according to the two specified nucleotides (which can be the same or different) to be recognized, select two corresponding single side unit vectors, and use NheI+HindIII to carry out double enzymes on the carrier that recognizes the 5' end base The carrier that recognizes the base at the 3' end is digested with SpeI+HindIII, and then the two DNA fragments containing the sequence of the side unit are connected to obtain a double side unit vector with the side unit arranged in series ( image 3 c in and image 3 in d). Select five basic single side unit carriers that recognize the four nucleotides of A, T, C, and G, and connect them in pairs. A total of 25 double side unit combinations can be obtained, which can identify all 16 possible dual cores. combination of nucleotide target sites. Using a similar...
Embodiment 3
[0053] Example 3 Using the "unit assembly" method to construct a paraunit tandem repeat vector that recognizes the target sequence of the zebrafish endogenous gene tnikb
[0054] Through the analysis, a target site located in the zebrafish gene tnikb was found, which was located at the junction of the first intron and the second exon of tnikb. Since the FokI cleavage domain needs to cut DNA in the form of a dimer, when using TALEN for gene targeting, it is necessary to design two TALE binding sites on the left and right, and the length of the spacer in the middle is generally 12~21bp. In this embodiment, the length of the left TALE binding site is 15bp, the right side is 16bp, and the interval between the two binding sites is 15bp ( Figure 6in a). The sequence of the left binding site is 5'-GTTATTTTCTCCCCT-3' (SEQ ID No.37). The steps of constructing the TALE repeat sequence combined with the above-mentioned sites are as follows: first step, use the double side unit carrie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com