Specific amplification of tumor specific DNA sequences
A tumor-specific and specific technology, applied in the direction of microbial determination/testing, biochemical equipment and methods, etc., can solve the problems of limited amplified loci, unlikely powerful methods for cancer detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1. Identification of tumor-associated hypomethylated regions
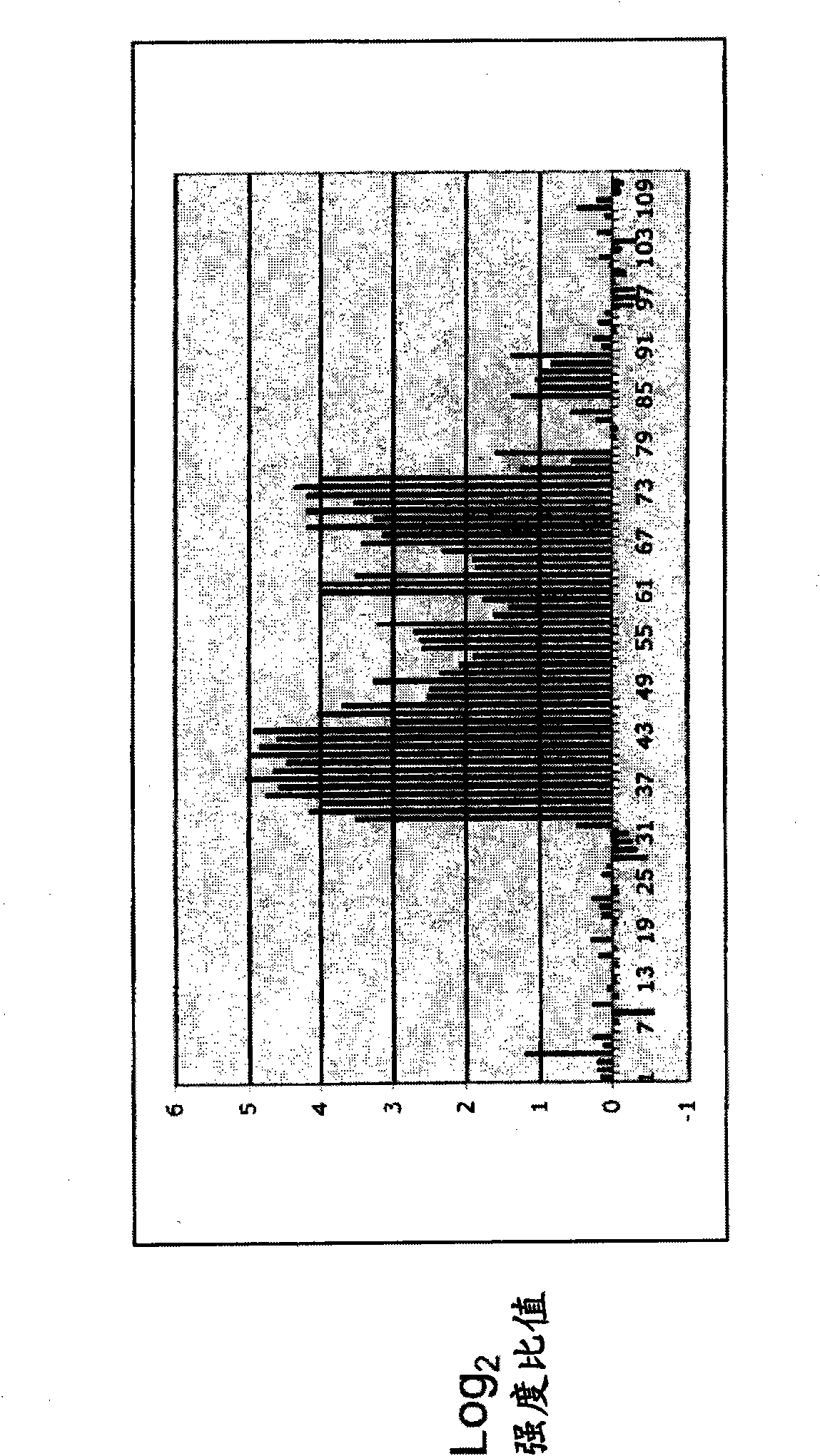
[0057] To test whether the CNA methylation profiles of subjects with ovarian tumors differed from those of normal controls, frozen serum samples were obtained from women whose blood was drawn prior to exploratory surgery due to suspicion of ovarian cancer. Similar samples were obtained from cancer-free women. DNA was prepared from 1 ml of cell-free serum by standard methods and all resulting samples were subjected to methylation-sensitive amplification as described above. Such a pair of samples was submitted to NimbleGen for hybridization to arrays that had previously been used to analyze trophoblast methylation.
[0058] The data indicated that both amplifications (cancer and normal) resulted in measurable signal (defined as >3 sd above background) from -5% of array addresses. Also, in ~70% of these cases, the log of the signal 2 - Ratios less than |1.5|, indicating that even though segments we...
Embodiment 2
[0060] Example 2. Development of Comparison Panel (Comparison Panel)
[0061] To facilitate detection and diagnosis using the methods of the invention, a normal population and a specific cancer patient population can be compared to develop a methylation profile associated with a specific type of cancer. The methods of the invention can be used to generate such methylation profiles.
[0062] DNA was isolated from serum or plasma of known cancer patients and normal controls using standard methods (Johnson, K.L. et al., Clin. Chem. 50:516-21 (2004)). Briefly, 10 ml of patient blood was centrifuged twice to remove cells. The resulting plasma was passed over a DNA-binding membrane. DNA was removed from the membrane and the resulting DNA was digested with HpyCh4-IV.
[0063] DNA adapters are annealed and ligated with digested DNA. The linker was designed to form a MluI restriction site when ligated with HpyCh4-IV digested DNA. Linker-mediated PCR was performed as described by...
Embodiment 3
[0068] Example 3. Preparation of microarrays for detection of hypomethylated tumor-associated DNA.
[0069] The genome was broken down into segments bounded by two sites for the relevant methylation-sensitive restriction enzyme (ACGT for HpyCh4-IV) and less than 500 base pairs in length. This provides a range of DNA segments that can be amplified from serum or plasma DNA samples. Algorithms are used to analyze the sequences of these fragments with the goal of finding suitable sequences for display on the microarray. For example, suitable oligonucleotides will have one or more of the following properties: (i) a unique sequence of greater than about 40 nucleotides, or a unique sequence of greater than about 60 nucleotides; (ii) about 40% to about 60% GC, and (iii) should not contain significant repeats or simple sequences, such as runs of greater than about 15 single bases. The array comprises oligonucleotides selected in this manner, wherein each oligonucleotide on the arra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com