Fusion gene microarray
A technology that combines genes and microarrays, applied in the determination/testing of microorganisms, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0112] Preparation of ligation probes (oligonucleotides) and microarrays
[0113] To generate ligation probes (oligonucleotides (oligos)), we produced computer scripts (written in the Python programming language) that automatically process public genomic data. We used the www.biomart.org Internet portal to obtain exon sequences for all genes and their transcripts. For each combination of fusion genes, the end sequence (last 30 nucleotides) of all gene A exons and the start sequence (30 nt) of all gene B exons were combined in all combinations. Next, an oligonucleotide calculation algorithm was used to generate probes (oligonucleotides) from these potential fusion gene exon-to-exon junctions. Here we optimize the Tm at 68°C and keep the Tm equal on each side of the junction. In our example, we generated exon-to-exon junction probes (oligonucleotides) that were 33-46 nucleotides in length.
[0114] In this way 47427 ligation probes (oligonucleotides) were designed for 275 fus...
Embodiment 2
[0119] functioning microarray
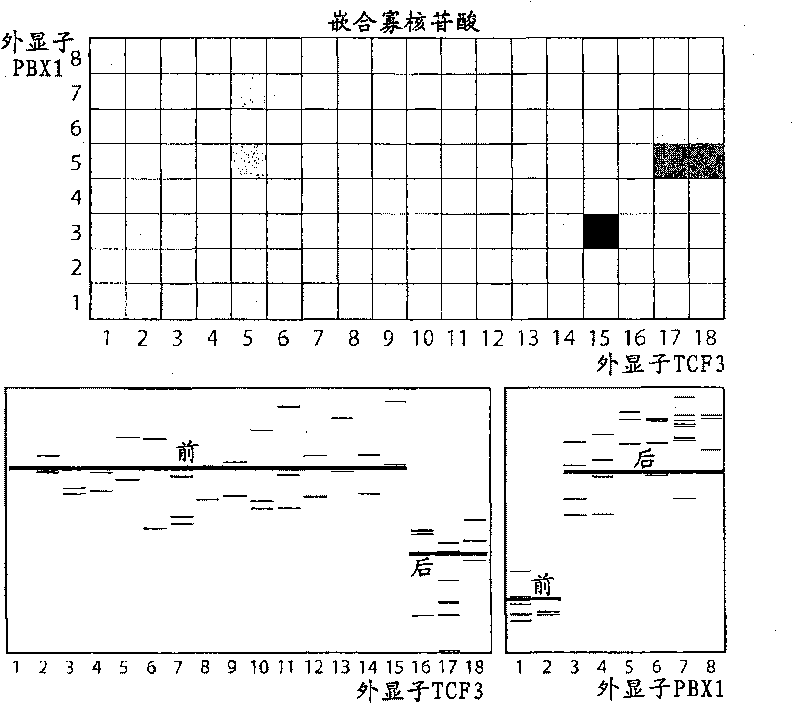
[0120] In a proof-of-principle experiment, we analyzed a set of positive control samples where each fusion gene was known to be present. Test samples included four prostate cancer tissue samples positive for the TMPRSS2:ERG fusion gene and two leukemia cell lines, each known to carry one of the TCF3:PBX1 fusion gene and the ETV6:RUNX1 fusion gene.
[0121] For test samples, total RNA was isolated by using a Qiagen spin column. In addition, ribosomal RNA reduction kit (RiboMinus TM Transcript Isolation Kit; Invitrogen) enriches for mRNA. From this, first-strand cDNA synthesis was performed with random primers (hexamers), followed by double-stranded cDNA and shipped to Nimblegen Inc. for labeling, hybridization, washing, and scanning of microarrays. cDNA was labeled by inclusion of Cy3- and Cy5-modified dNTPs.
[0122] in conclusion
[0123] To visualize the measurement of the positive control gene, we performed two separate pathways using ch...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com