Protein related to rice ear sprouting period and encoding genes and uses thereof
A technology that encodes genes and heading dates is applied in the fields of application, genetic engineering, and plant genetic improvement to achieve the effects of expanding crop planting area, early maturity, and increasing crop yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1, the acquisition of OsDof12 and its coding gene
[0050] 1. Acquisition of the cDNA sequence of a protein related to rice heading stage
[0051] The rice variety Nipponbare was used as the experimental material, and total RNA was extracted from its leaves, which was reverse transcribed into cDNA. Using this cDNA as a template and using 5'-gtgcgaatgaaaagatttcaag-3' and 5'-gatctaaatattcaaatcattatatt-3' as primers, the cDNA sequence of a protein related to rice heading stage was amplified by PCR. The reaction mixture is as follows:
[0052] wxya 2 o
18.3μl
10×PCR buffer
2.5μl
dNTP Mixture (2.5mM)
2.5μl
Taq enzyme (5U / μl)
0.2μl
Primer (10mM)
0.25μl
Primer (10mM)
0.25μl
Template (reverse transcribed cDNA)
1μl
Total
25μl
[0053] PCR reaction conditions: pre-denaturation at 94°C for 4min; then denaturation at 94°C for 45S; annealing at 55°C for 45S; extension at 7...
Embodiment 2
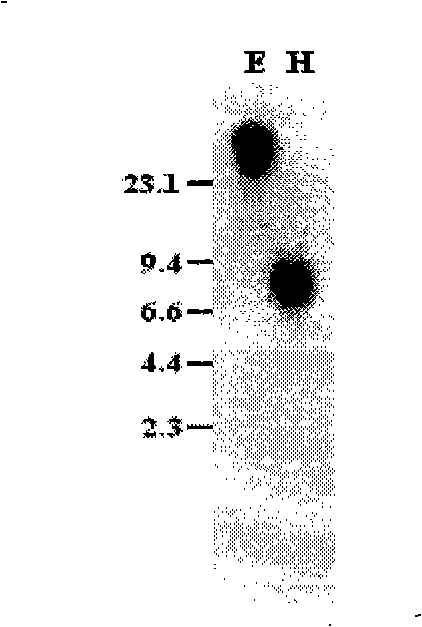
[0062] Embodiment 2, Southern blot analysis
[0063] Genomic DNA of rice variety Nipponbare was extracted, digested with restriction endonucleases EcoR I and Hind III respectively and transferred to the membrane, using the OsDof12 full-length cDNA sequence obtained in the above example 1 from the 670th to 1378th deoxyribose nucleus at the 5' end The nucleotide fragments were used as probes for hybridization analysis. Hybridization results such as figure 2 As shown, where E is the result of Southern blot after EcoRI single-enzyme digestion, and H is the result of Southern blot after Hind III single-enzyme digestion. From figure 2 It can be seen that the hybridization signals of EcoR I and Hind III single-enzyme digestion are all single bands, indicating that OsDof12 exists in the form of a single copy in the rice genome.
Embodiment 3
[0064] Example 3. Analysis of subcellular localization and transcriptional activation domain of OsDof12
[0065] 1. Analysis of subcellular localization of OsDof12
[0066] As a result of analyzing the OsDof12 protein obtained in Example 1 above, no sequence similar to the reported nuclear localization signal was found. In order to analyze the localization of OsDof12 in cells, OsDof12 was digested with Nco I and the GFP-containing vector CaMV35S-sGFP(S65T)-nos3'(pUC18) (see reference Niwa Y , Hirano T, Yoshimoto K, Shimizu M, Kobayashi H.Non-invasivequantitative detection and applications of non-toxic, S65T-type green fluorescent proteinin living plants.Plant J.199918 (4): 445-463.) connected to get Recombinant expression vector 35S-OsDof12-GFP. After no frameshift was verified by sequencing, the recombinant expression vector 35S-OsDof12-GFP was transferred into onion epidermal cells with a gene gun, and the onion epidermal cells transformed with the empty vector CaMV35S-sGF...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com