Hcv ns3-ns4a protease inhibition
一种NS3-NS4A、蛋白酶的技术,应用在治疗HCV感染,抑制丙型肝炎病毒NS3-NS4A蛋白酶活性的化合物领域,能够解决抗HCV疫苗前景不明朗等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
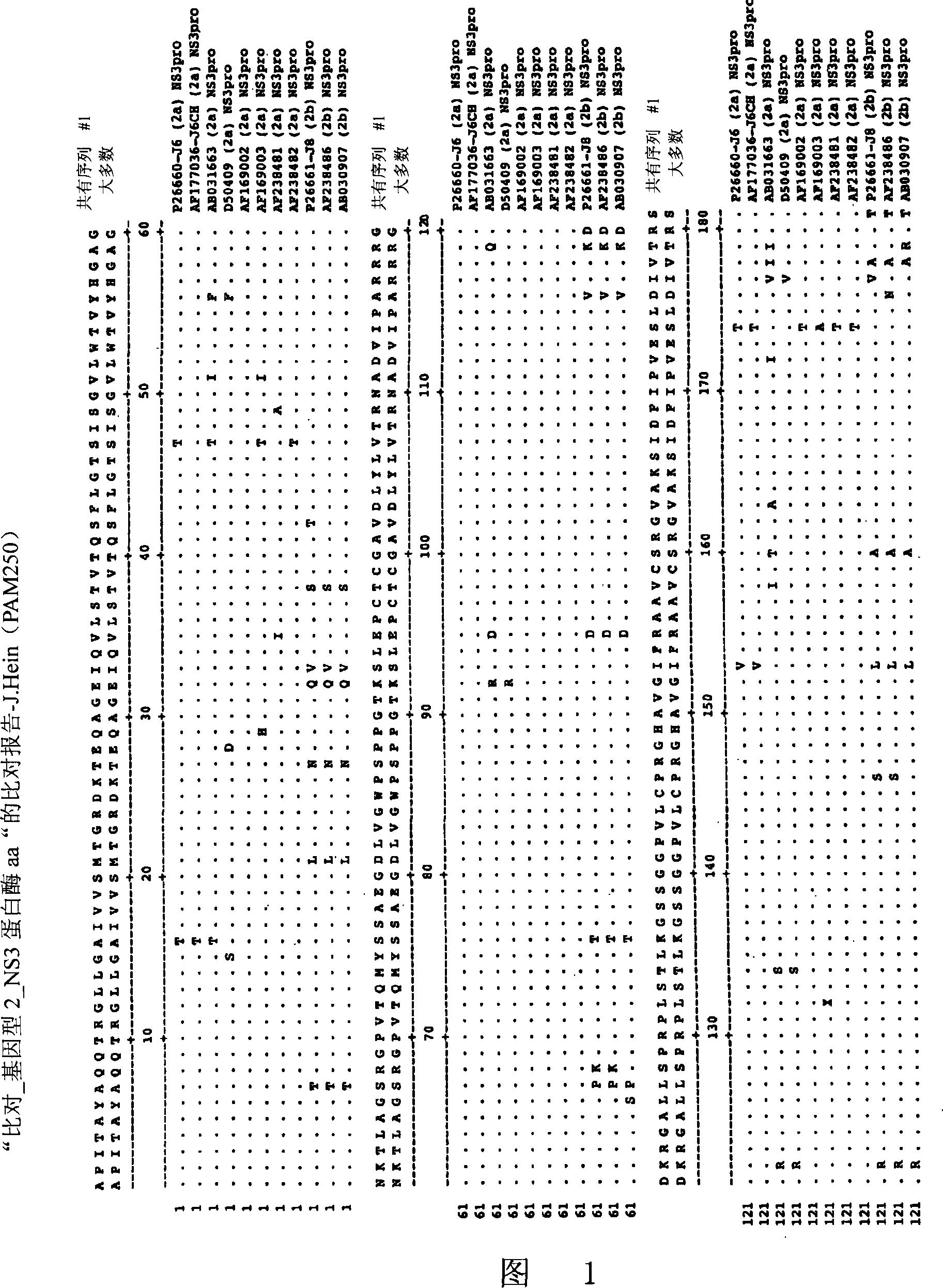
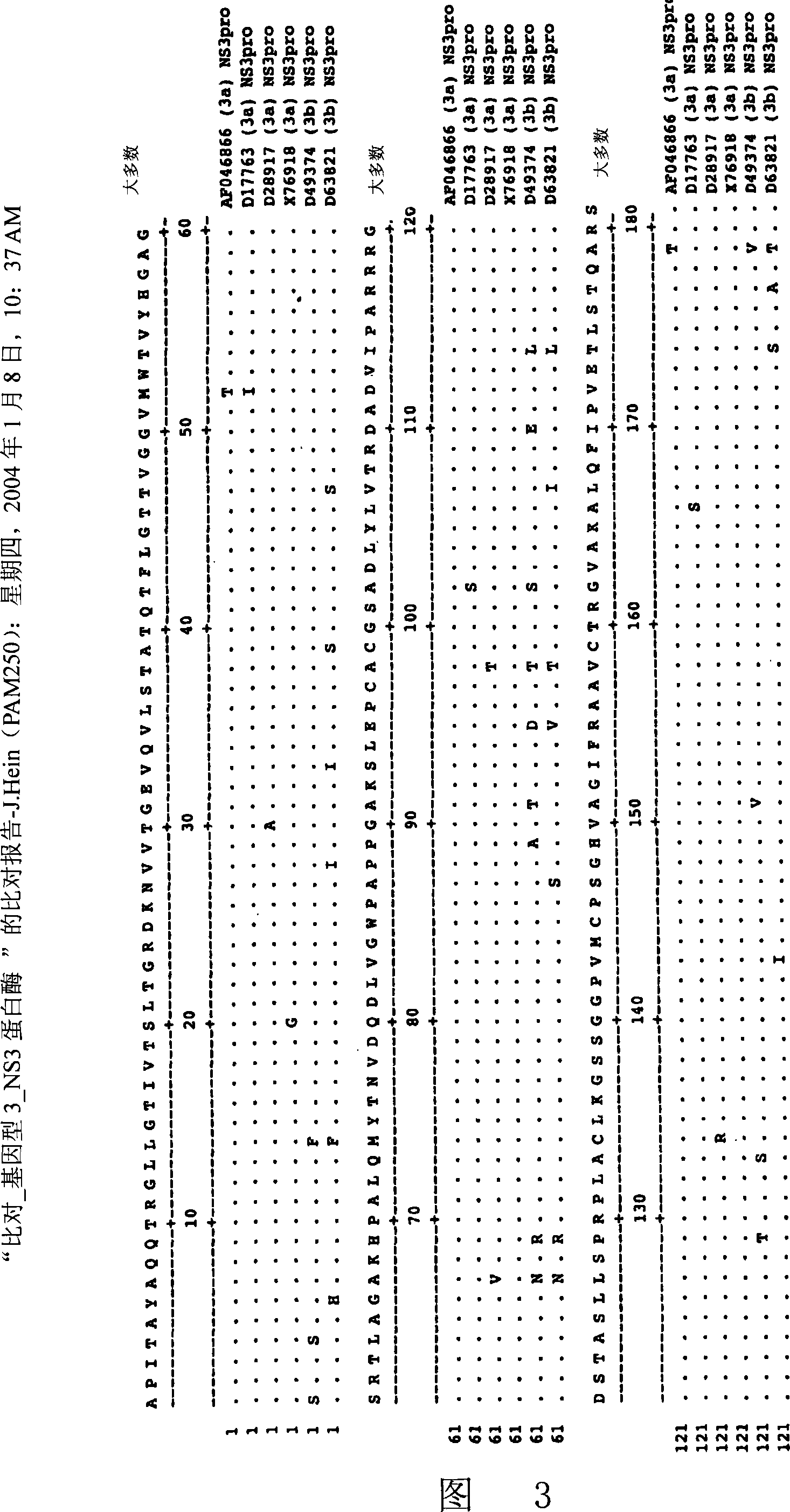
Image
Examples
Embodiment 1
[0107] HCV NS3 Protease HPLC Peptide Cleavage Assay
[0108] This method is a modification of the method described by Landro et al. [Landro JA et al. Biochemistry, 1997, 36:9340-9348]. For all proteases, a peptide substrate based on the NS5A / NS5B split site of HCV genotype la (NS5AB) was used. 25 mM substrate stocks were prepared in DMSO containing 0.2 M DTT and stored at -20°C. The central core region of NS4A was replaced by a synthetic peptide cofactor (KK4A) appropriate for each genotype. See below for peptide sequences. Hydrolysis reactions were performed on 96-well microtiter plates with 25nM-50nM HCV NS3 protease in a buffer containing 50mM HEPES (pH 7.8), 100mM NaCl, 20% glycerol, 5mM DTT and 25μM KK4A. The final concentration of DMSO was no more than 2% (v / v). The reaction was terminated by adding 10% trifluoroacetic acid (TFA) to make the final concentration of TFA 2.5%. Substrate and product were separated on a reverse-phase microbore HPLC column (Phenomenex Jup...
Embodiment 2
[0113] Potency Determination by HPLC Peptide Fractionation
[0114] In order to determine the apparent Ki value, all components except the test compound and substrate were pre-incubated at room temperature for 5 min. Then the test compound dissolved in DMSO was added to the mixture and incubated at room temperature for 15 min. Add NS5AB peptide at a concentration equal to Km (11 μM-70 μM), and incubate at 30° C. for 15 min to initiate the reaction. Enzyme inhibition activity was titrated with 7 to 8 concentrations of compound. Via non-linear regression, the activity and corresponding inhibitor concentration data points were fitted to the Morrison equation [SculleyMJ & Morrison JF, Biochim Biophys Acta, 1986, 874: 44-53] describing the inhibition reaction of competitive strong binding enzymes.
[0115] VX-950 pairs measured by peptide cleavage
Embodiment 3
[0117] HCV NS3 Protease Fluorescent Peptide Assay
[0118]Enzyme activity was determined by a modification of the method described by Taliani et al [Taliani M et al. Anal Biochem, 1997, 240:66-67]. All reactions used RET-S1 fluorescent peptide (AanSpec, San Jose, CA) as substrate in buffer A (buffer containing 50 mM HEPES (pH 7.8), 100 mM NaCl, 20% glycerol, 5 mM DTT and 25 μM KK4A) conduct. The final concentration of DMSO was maintained at 1-2% (v / v). Unless otherwise stated, reactions were monitored serially using a fluorescent microtiter plate reader thermostatted at 30°C with excitation and emission filters of 355 nm and 495 nm, respectively.
[0119] In order to determine the kinetic parameters Km and Vmax, the concentration of substrate RET-S1 in buffer A was varied between 6 μM and 200 μM, and it was reacted with 5 nM~10 nM HCV NS3 protease for 5~10 min. Add 25 μL of 10% TFA to stop the reaction. The substrate and product were separated on a reverse-phase microbore ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com