Pathway recognition algorithm using data integration on genomic models (paradigm)
a path recognition and genomic model technology, applied in the field of computational biology, can solve the problems of spia being generally limited to using only a single type of genome-wide data, spia's analytic and predictive value remains highly restricted, and all or almost all of the currently known pathway analyses fail, etc., to achieve the effect of facilitating presentation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
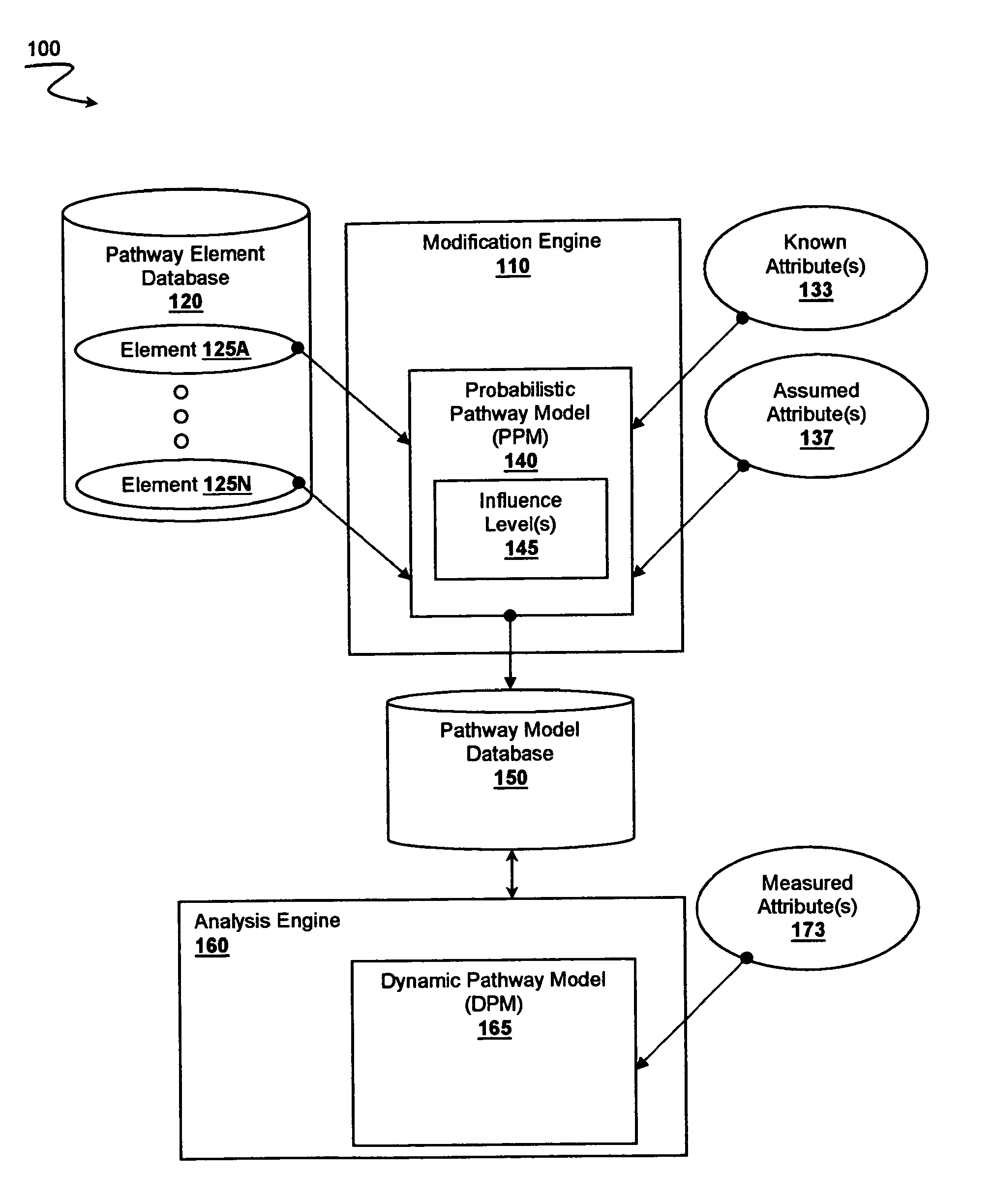
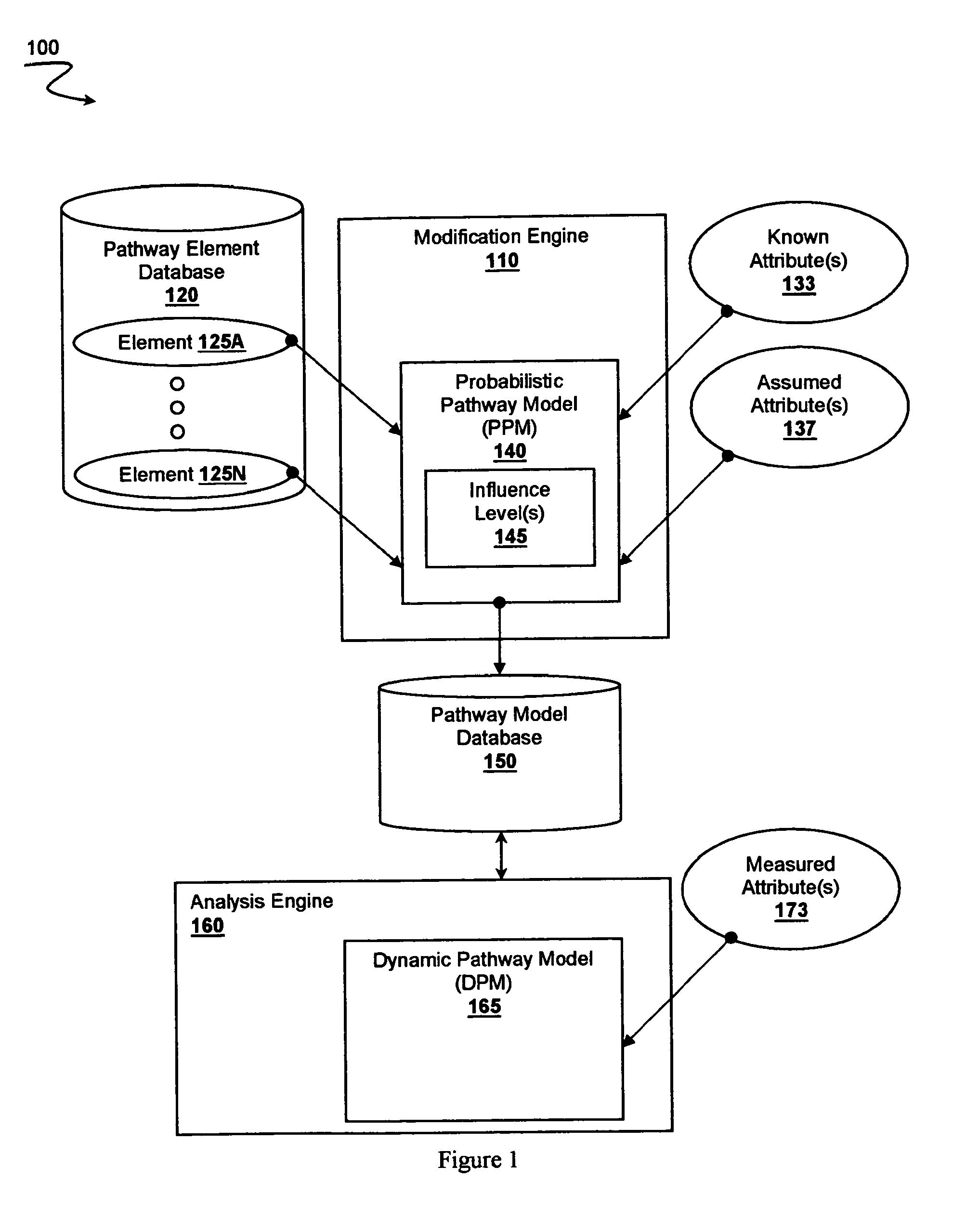
[0020]The inventors have developed systems and methods where multiple attributes of multiple pathway elements are integrated into a probabilistic pathway model that is then modified using patient data to produce a dynamic pathway map. Most significantly, it should be appreciated that the attributes for pathway elements within a pathway need not be known a priori. Indeed, at least some of the attributes of at least some pathway elements are assumed. The pathway elements are then cross-correlated and assigned specific influence levels on or more pathways to so construct the probabilistic pathway model, which is preferably representative of a particular reference state (e.g., healthy or diseased). Measured attributes for multiple elements of a patient sample are then used in conjunction with the probabilistic pathway model to so produce a patient sample specific dynamic pathway map that provides reference pathway activity information for one or more particular pathways.
[0021]It should ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com