Pullulanase Variants and Uses Thereof
a technology of pullulanase and variants, which is applied in the field of pullulanase variants, can solve the problems of unfavorable panose formation in the saccharification process, and achieve the effect of high stringency conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0163]Cloning and Expression of Thermococcus hydrothermalis DSM 14834 Pullulanase in Bacillus subtilis
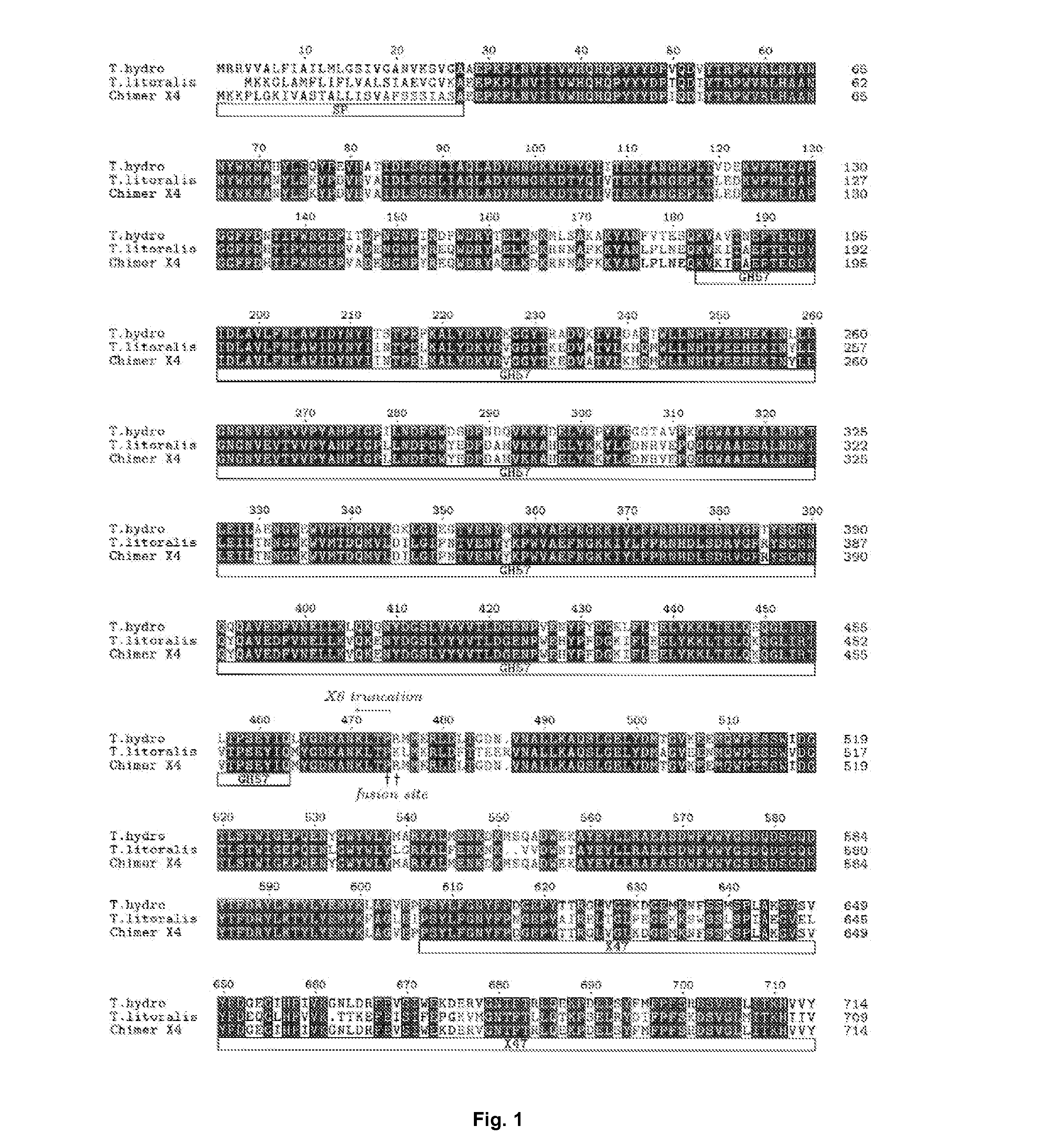
[0164]A synthetic gene based on the protein sequence of Thermococcus hydrothermalis DSM 14834 Apu (Uniprot: Q9Y8I8) was designed and the gene was codon optimized for Bacillus subtilis. The C-terminal sequence (including the part of the putative linker, indicated in the alignment—FIG. 1) TPTESPTETTTTTPSETTTTTSTTTGPSSTTTSTPGGGICGPGIIAGLALIPLLLKRRN (SEQ ID NO: 5) and the native signal peptide (MRRVVALFIAILMLGSIVGANVKSVG—SEQ ID NO: 6) was not part of the designed synthetic gene (SEQ ID NO: 1 (DNA) and SEQ ID NO: 2 (protein)).The synthetic gene was extracted by double-digestion according to the manufacturer's manual (FastDigest®, Fermentas, Germany) from a plasmid carrying the synthetic gene. The synthetic gene was cloned in an expression vector comprising the genetic elements as described in WO 99 / 43835 (hereby incorporated by reference). By doing so, the signal peptide from the alkali...
example 2
Cloning and Expression of Variants of Thermococcus hydrothermalis DSM 14834 Pullulanase in Bacillus subtilis
[0167]In order to increase the recombinant expression yield of Apu and to identify which domains are necessary for the pullulanase activity, we have made truncations of the enzyme. The expression plasmid containing the synthetic gene described in Example 1 was used as template for a PCR(1). PCR(1) was performed in a total volume of 26 microliters, the following reagents were added, 1 microliter of vector DNA preparation (template), 50 pmol of each of the primer pairs below, 5 microliters dNTPs and 0.5 microliter Phusion® polymerase (Finnzymes, Finland) in Phusion HF buffer. The PCR conditions were 98° C. for 2 min; 9 cycles of 98° C. for 15 sec; 65° C. for 45 sec; 72° C. for 4 min; followed by 72° C. for 10 min; 4° C. for 20 min and 15° C. until the end of the PCR program. The primer pairs used were:
For X4 truncation:
For X4 truncation:1354(SEQ ID NO: 31)5′- GCCAAGGCCGGTTTTTTA...
example 3
[0170]Cloning and Expression of a Truncated Pullulanase Fusion of Thermococcus litoralis DSM 5473 and Thermococcus hydrothermalis DSM 14834 in Bacillus subtilis
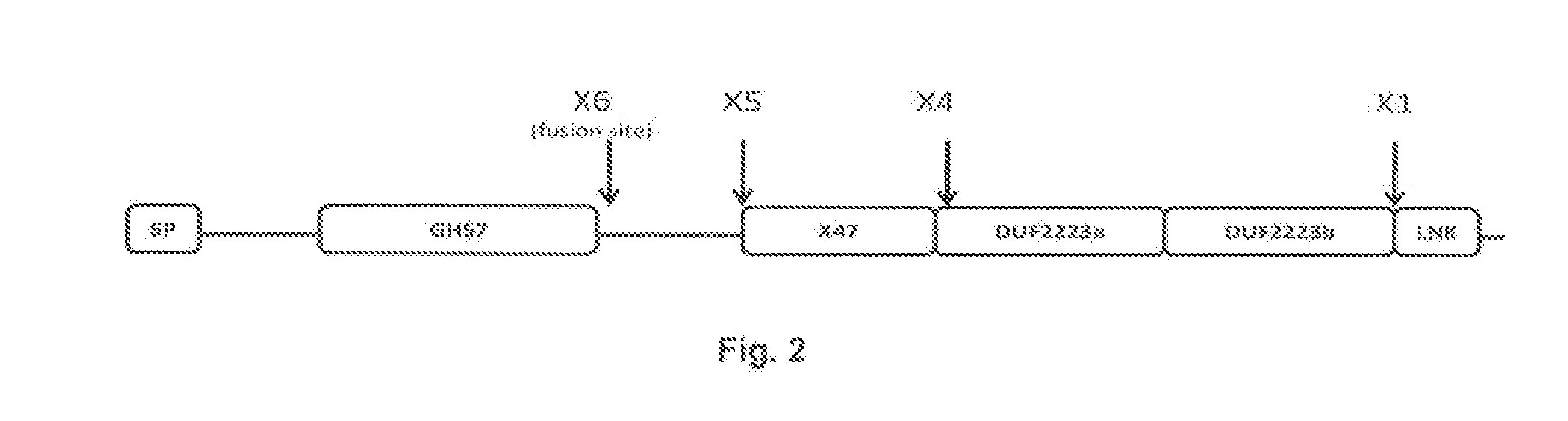
[0171]A truncated synthetic gene based on the protein sequence of Thermococcus litoralis DSM 5473 was designed and the gene was codon optimized for Bacillus subtilis. In the synthetic gene, the C-terminal sequence after amino acid 474 in SEQ ID NO: 4 was deleted as indicated in the alignment FIG. 2 (“X6 truncation”).
[0172]Cloning of the synthetic gene into Bacillis subtilis the selection of correct clones and the cultivation of the clones was performed as described in Example 1. The X6 truncation of T. litoralis did not display any pullulanase activity (see table in Example 4). The derived expression plasmid C526A was used in a following PCR amplification as template.
Fusion of Thermococcus litoralis and Thermococcus hydrothermalis DNA
[0173]Using the expression plasmid C526A carrying the apu gene from Thermococcus litoralis a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com