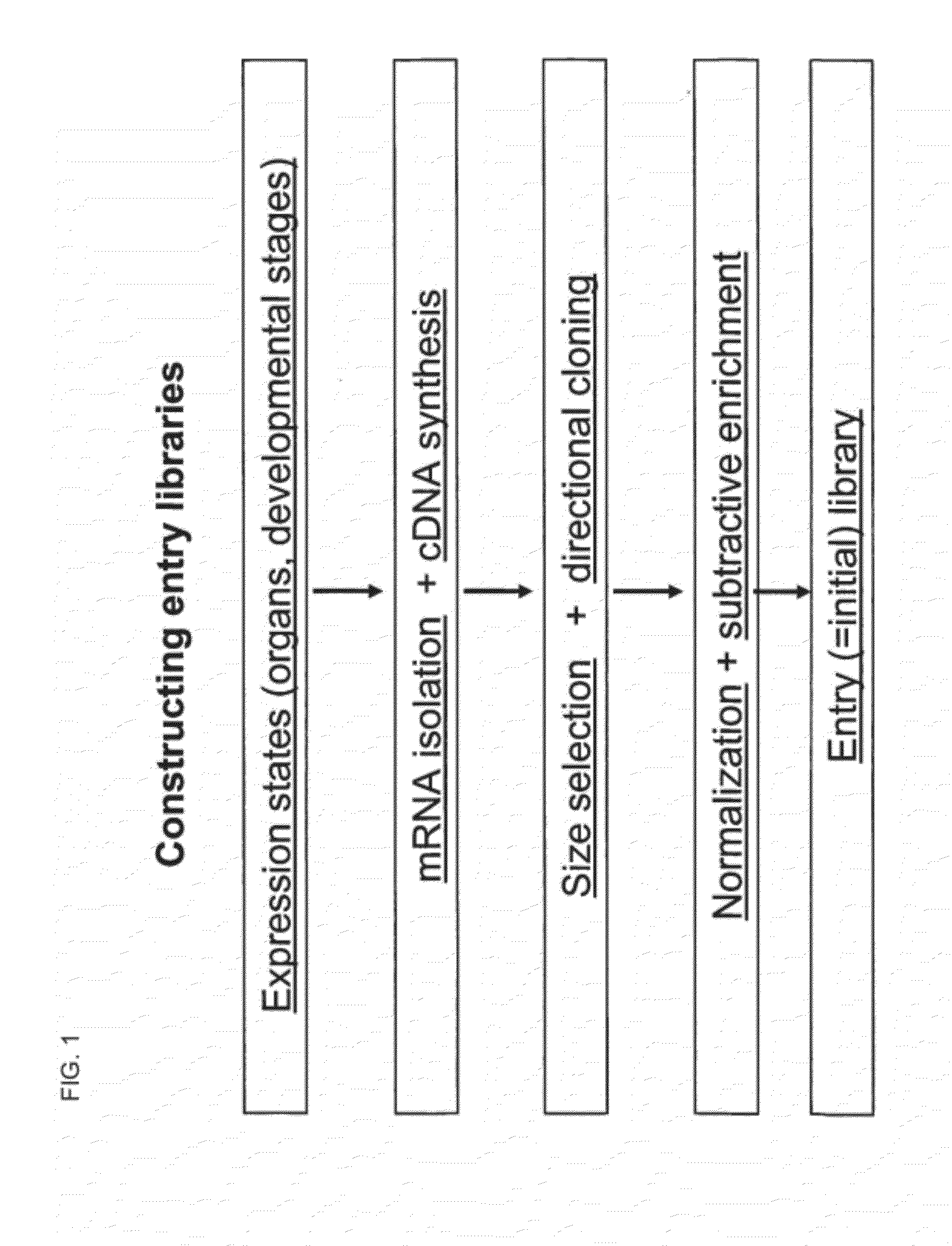
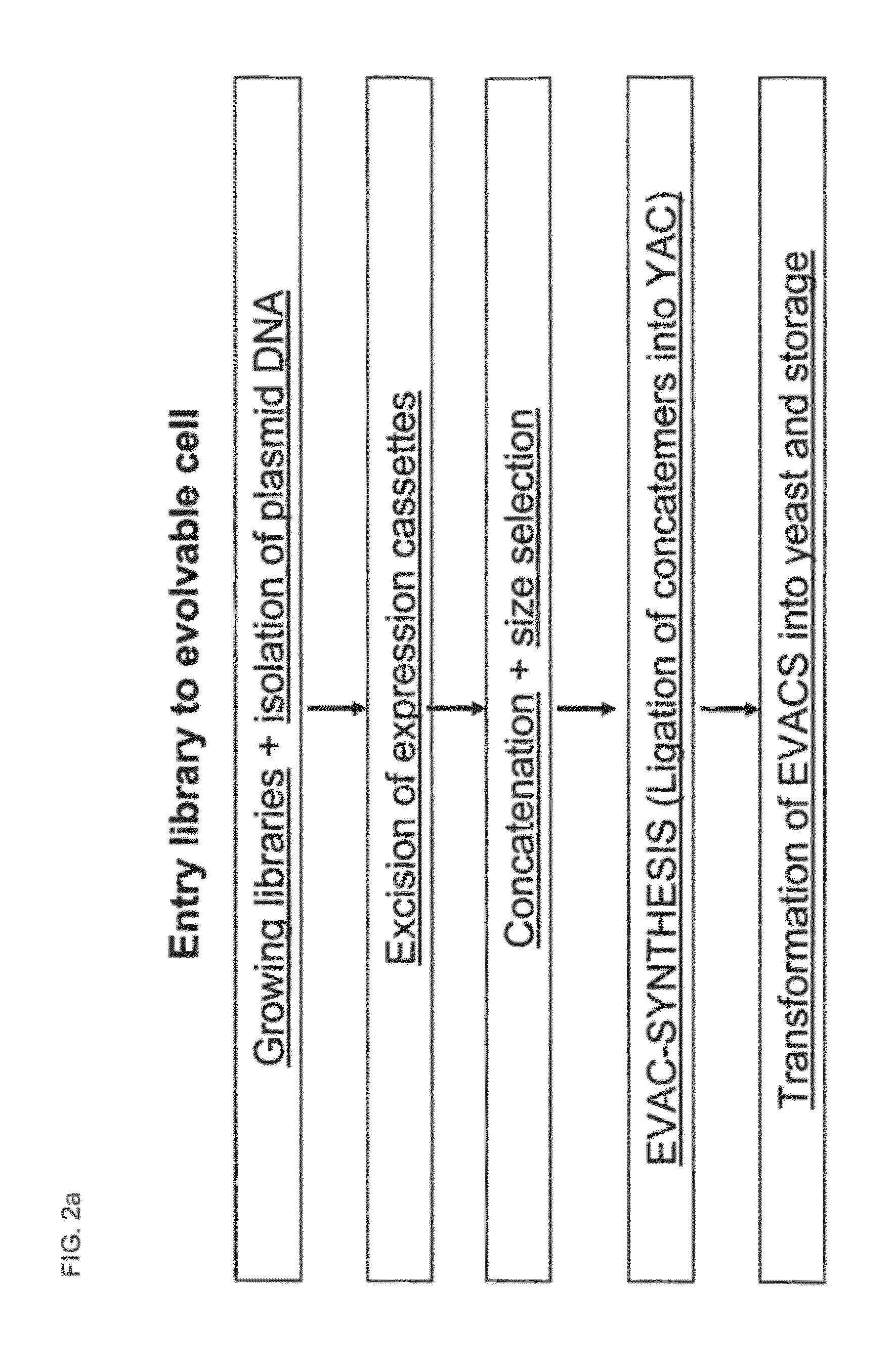
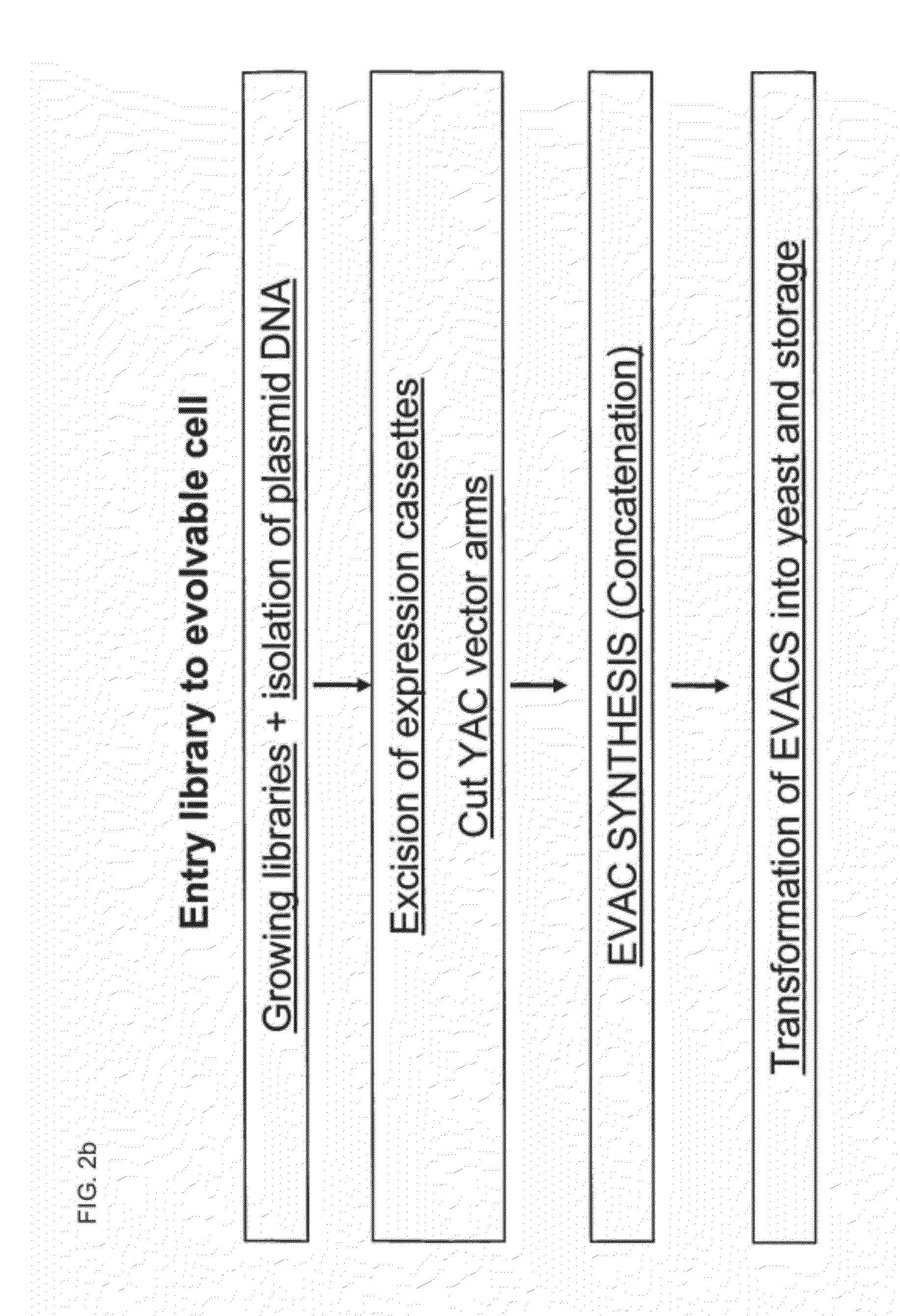
Library of a Collection of Cells
a cell collection and gene technology, applied in the field of combinatorial gene expression libraries, can solve the problems of large chance, heterologous genes are lethal or sub-lethal to the cell,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0313]In the examples 1-3 an AscI site was introduced into the EcoR1 site in pYAC4 (Sigma, Burke D T et al. 1987, Science vol 236, p 806), so that sticky ends match the Asc1 site(=RS2 in general formula of this patent) of the cassettes in pEVE vectors
Preparation of EVACS (Evolvable Artificial Chromosomes) Including Size Fractioning
[0314]Preparation of pYAG4-Asc Arms
1. inoculate 150 ml of LB (sigma) with a single colony of E. coli DH5α containing pYAC4-Asc
2. grow to OD600˜1, harvest cells and make plasmid preparation
3. digest 100 μg pYAC4-Asc w, BamH1 and Asc1
4. dephosphorylate fragments and heat inactivate phosphatase (20 min, 80 C)
5. purify fragments(e.g. Qiaquick Gel Extraction Kit)
6. run 1% agarose gel to estimate amount of fragment
Preparation of Expression Cassettes
[0315]1. take 100 μg of plasmid preparation from each of the following libraries
a) pMA-CAR
b) pCA-CAR
c) Phaffia cDNA library
d) Carrot-cDNA library
2. digest w. Srf1(10 units / prep, 37 C overnight)
3. dephosphorylate (10 u...
example 2
Preparation of EVACs (Evolvable Artificial Chromosomes) with Direct Transformation
[0335]Preparation of pYAC4-Asc Arms
1. inoculate 150 ml of LB with a single colony of DH5α containing pYAC4-Asc
2. grow to OD600˜1, harvest cells and make plasmid preparation
3. digest 100 μg pYAC4-Asc w. BamH1 and Asc1
4. dephosphorylate fragments and heat inactivate phosphatase(20 Min, 80 C)
5. purify fragments(e.g. Qiaquick Gel Extraction Kit).
1. run 1% agarose gel to estimate amount of fragment
Preparation of Expression Cassettes
[0336]1. take 100 μg of plasmid preparation from each of the following libraries
e) pMA-CAR
f) pCA-CAR
g) Phaffia cDNA library
h) Carrot cDNA library
2: digest w. Srf1 (10 units / prep, 37 C overnight)
3. dephosphorylate (10 units / prep, 37 C, 2 h)
4. heat inactivate 80 C, 20 min
5. concentrate and change buffer (precipitation or ultra filtration),
6. digest w. Asc1. (10 units / prep, 37 C, overnight)
7. adjust volume of preps to 100 μL
Preparation of EVACs
[0337]Different types of EVACs have bee...
example 3
Preparation of EVACs (EVolvable Artificial Chromosomes) (Small Scale Preparation)
Preparation of Expression Cassettes
[0355]1. inoculate 5 ml of LB-medium (Sigma) with library inoculum corresponding to a 10+fold representation of library. Grow overnight[0356]2. make plasmid miniprep from 1.5 ml of culture (E.g. Qiaprep spin miniprep kit)[0357]3. digest plasmid w. Sri 1[0358]4. dephosphorylate fragments and heat inactivate phosphatase (20 min, 80 C)[0359]5. digest w. Asc1[0360]6. run 1 / 10 of reaction in 1% agarose to estimate amount of fragment
Preparation of pYAC4-Asc Arms[0361]1. inoculate 150 ml of LB with a single colony of E. coli DH5α containing pYAC4-Asc[0362]2. grow to OD600˜1, harvest cells and make plasmid preparation[0363]3. digest 100 μg pYAC4-Asc w. BamH1 and Asc1[0364]4. dephosphorylate fragments and heat inactivate phosphatase (20 min, 80 C)[0365]5. purify fragments(E.g. Qiaquick Gel Extraction Kit)[0366]6. run 1% agarose gel to estimate amount of fragment
Preparation of E...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| salinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com