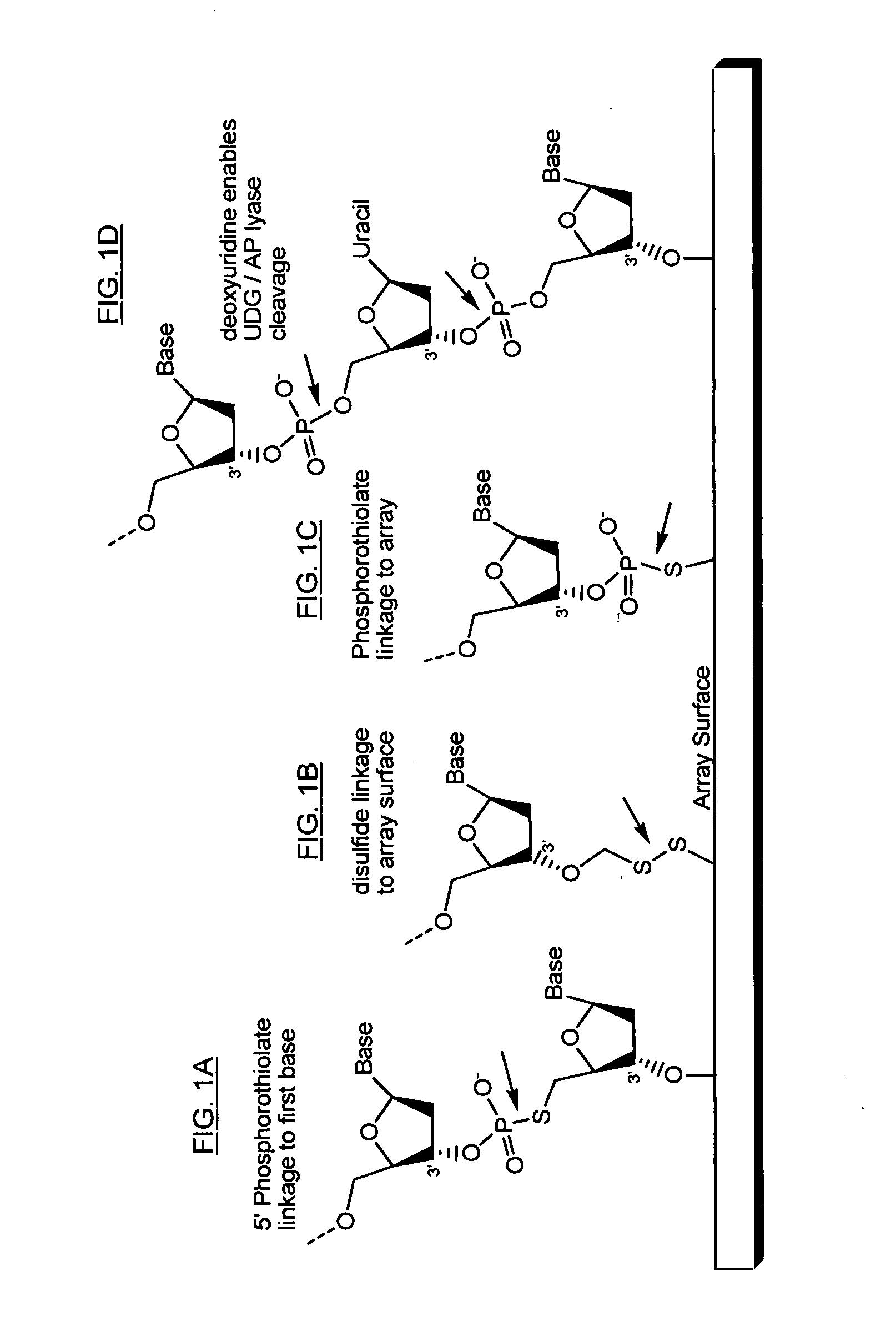
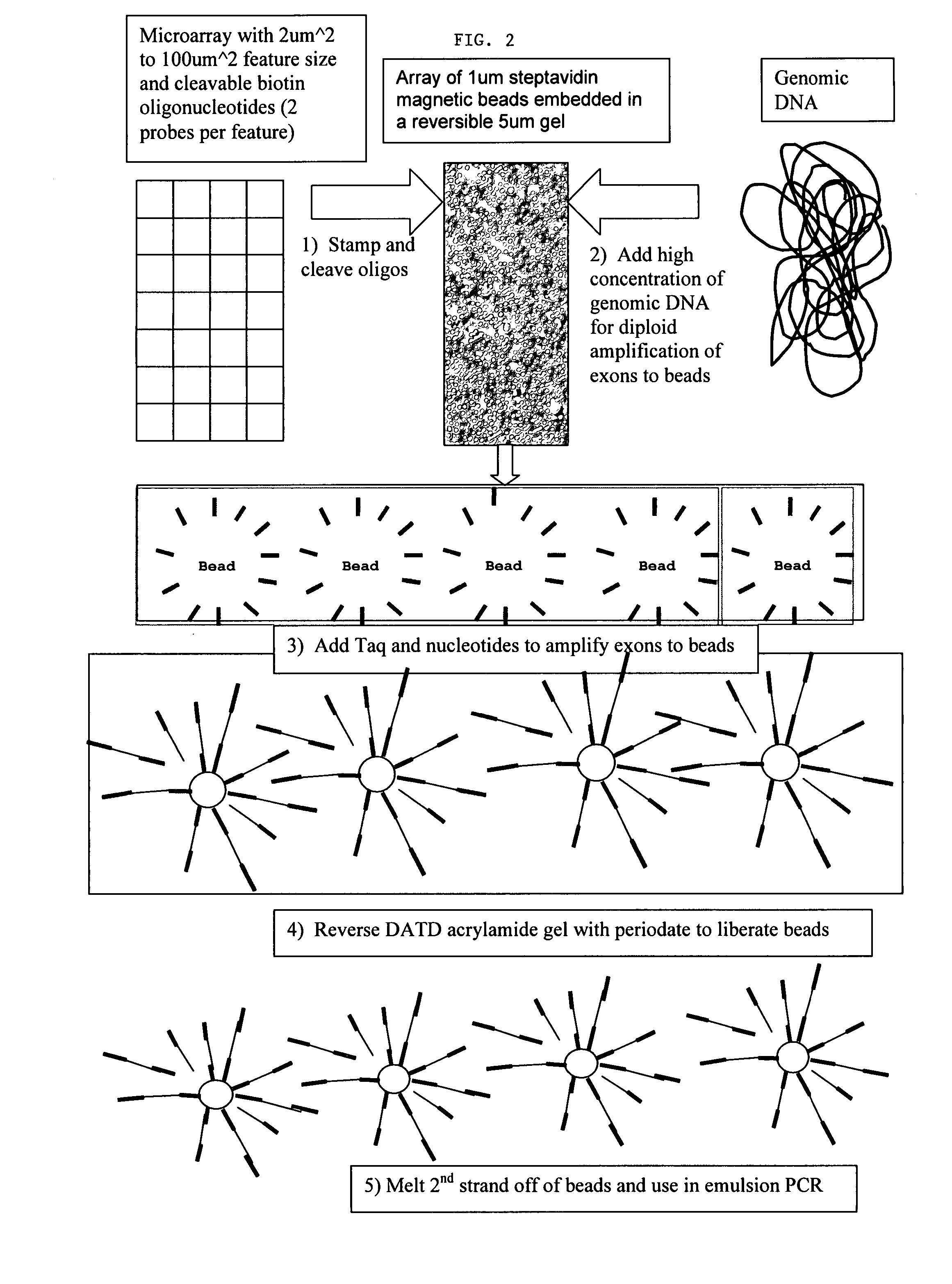
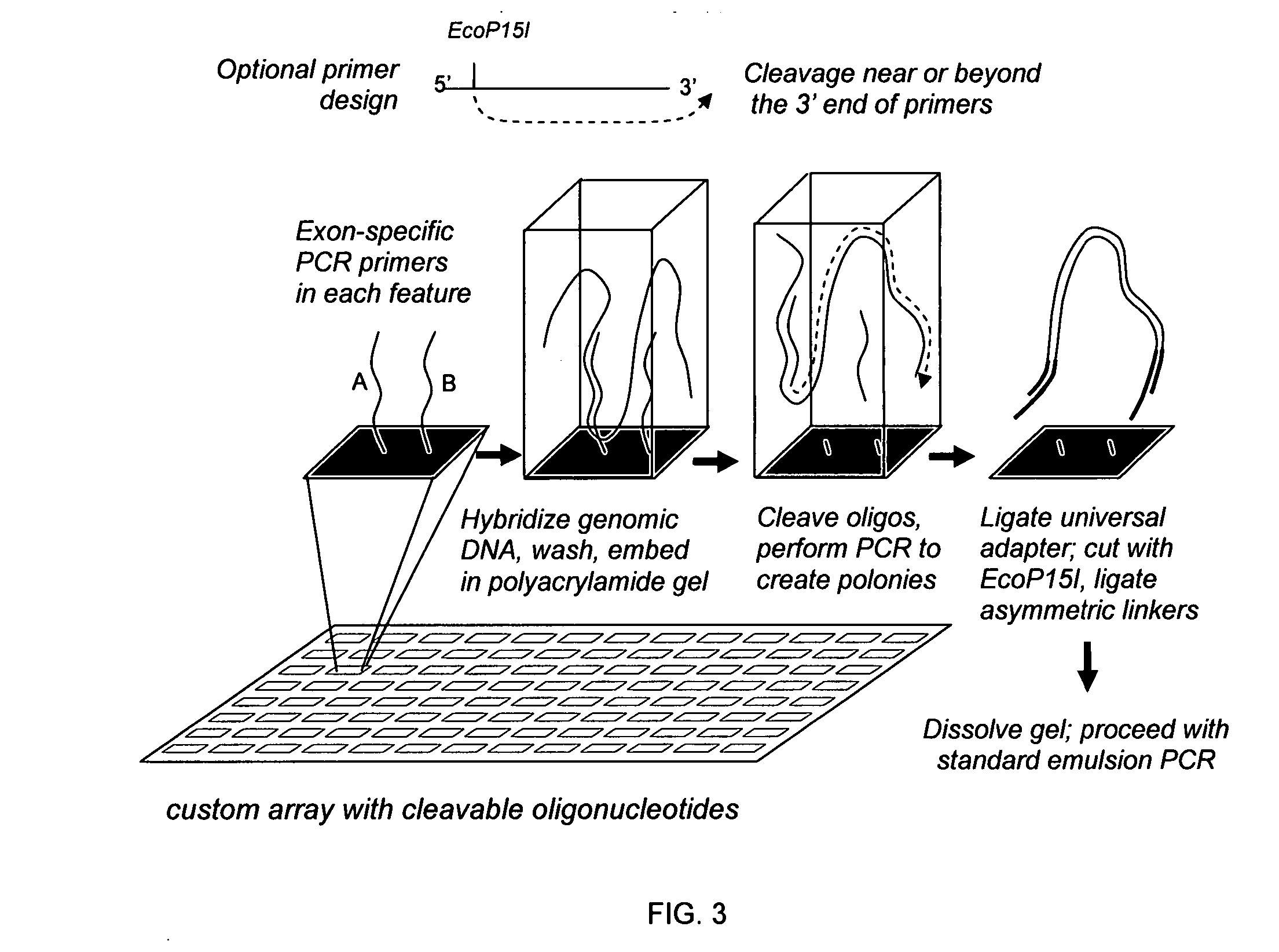
Directed enrichment of genomic DNA for high-throughput sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0061] Amplifying all of the exons in the human genome requires 250,000 amplicons or polymerase chain reaction (PCR) products. Manufacturing the required 250,000 primer pairs is a challenging task. Microarray technologies offer an attractive approach for the synthesis of very small quantities of up to 400,000 oligonucleotides on a single solid support. However, liberating or releasing these 400,000 oligonucleotides from the microarray for use as amplification primers (e.g., for PCR) has not been useful because there is no current method for segregating the appropriate primer pairs into individual amplification reactions. It is known that all 400,000 oligonucleotides can be released into a single PCR amplification reaction. But, the resulting multiplex PCR reaction has little informative value due to the complexity of the amplified products generated and due to numerous artifacts that result from such a multiplex PCR, such as primer-dimer formation. Thus, there is a need for methods ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Area | aaaaa | aaaaa |
| Area | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com