Methods and means for obtaining modified phenotypes
By introducing unpolyadenylated RNA molecules with target-specific sequences through chimeric DNA in plant cells, the method effectively addresses inefficiencies in gene silencing in transgenic plants, achieving robust reduction of nucleic acid expression and improved phenotypic control.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Experimental Procedures
1.1 Chimeric DNA Constructs
Ribozyme-Containing GUS Gene Constructs and a Control Construct
[0141] The ribozyme sequences used are the plus strand or negative strand self-cleavage sequences of the satellite RNA of the barley yellow dwarf virus (BYDV) RPV serotype, which was isolated in CSIRO Plant Industry (SEQ ID 1 and 2; Miller et al., 1991).
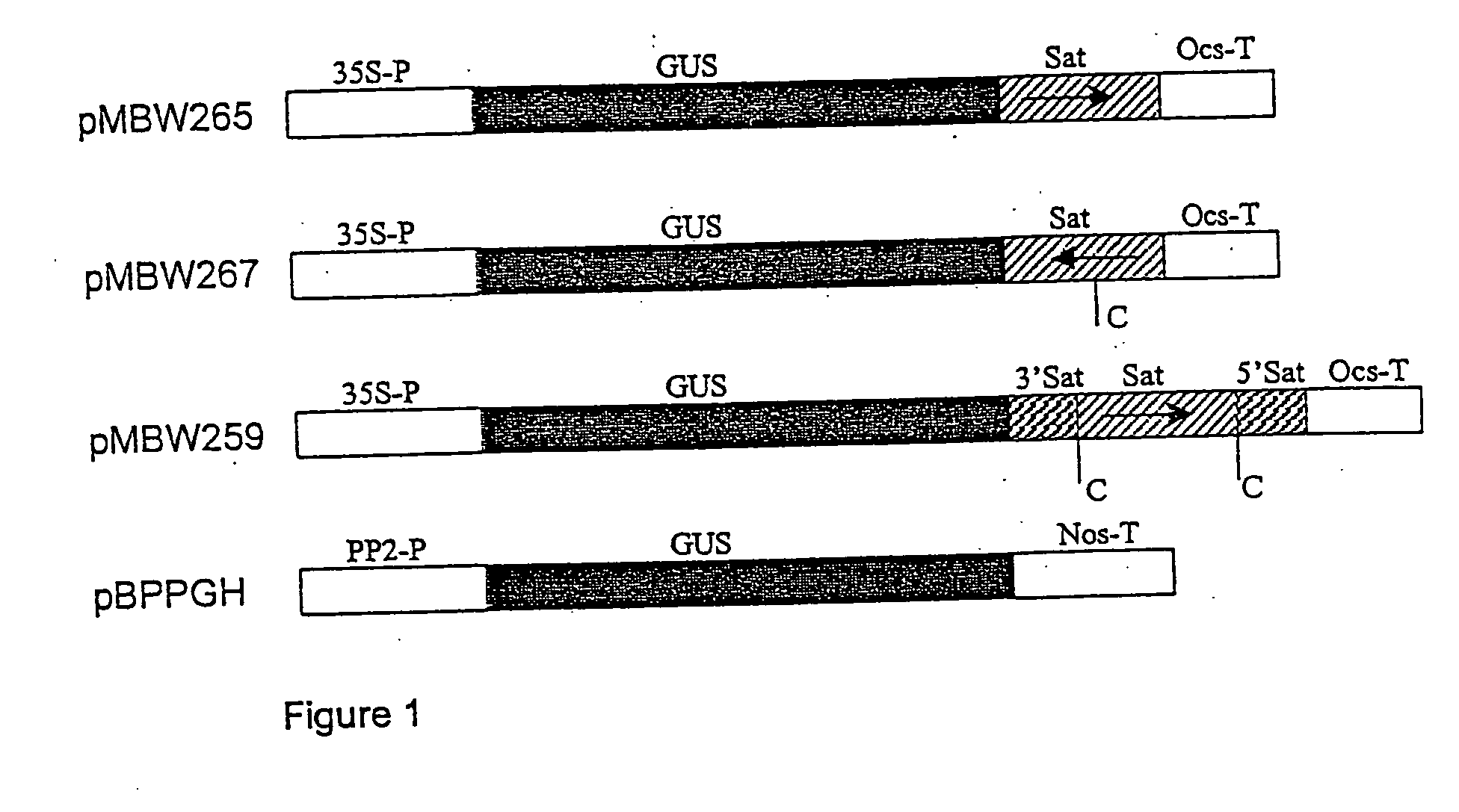
[0142] The two ribozyme-containing GUS constructs (pMBW259 and pMBW267) and one control GUS construct (pMBW265) are schematically drawn in FIG. 1. pMBW259 contains two plus strand cleavage sites, while pMBW267 contains the negative strand cleavage site.
[0143] To make these constructs, β-glucuronidase (GUS) gene sequence was modified to contain a NcoI site around the translational start ATG and cloned into pART7 (Gleave, 1992) at the XhoI / EcoRI sites, forming pMBW258. The full-length BYDV-RPV satellite sequence was amplified by PCR using primers SatPR1 (SEQ ID No. 3) and SatPR4 (SEQ ID No. 6), digested with BamHI and...
example 2
GUS Expression in Transgenic Tobacco Transformed with a Single Type of the GUS Constructs
[0148] Transgenic plants containing pMBW259 and pMBW267 showed very low levels of GUS expression, as judged by lack of, or faint blue, GUS staining. Plants transformed with pMBW265 showed more GUS expression than with pMBW259 and pMBW267, but the level was much lower than plants transformed with pBPPGH. The best pMBW265 lines expressed 13.3% of the GUS activity by an average pBPPGH line.
example 3
Gus Expression in Super-Transformed Lines Containing pBPPGH and One of the Three Other Constructs of Example 1
[0149] In order to promote silencing of a normal GUS gene by the presence of the ribozyme sequence near the 3′ end of the GUS gene transcript, plants containing pMBW259, pMBW265 or pMBW267 and pBPPGH were constructed by re-transformation. Histochemical GUS assays of the super-transformants showed that the pMBW267 background gave substantially higher proportions of transformants than the pMBW259 or the pMBW265 background that showed low levels of GUS expression as indicated by the lack of strong and uniform blue staining. Super-transformants containing pBPPGH and pMBW265 showed the best GUS expression.
[0150] Table 2 shows the result of fluorometric GUS (MUG) assay of the super-transformants. The lines (E and F) containing pBPPGH and pMBW267 showed uniformly low GUS expression compared with the other lines. The best GUS expression came from the C lines which contain pBPPGH a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com