Immunoglobulin variants and uses thereof
a technology of immunoglobulin and variants, applied in the field of can solve the problem of limit the use of murine antibodies in human therapy, human anti-mouse antibody (hama) response, etc., to improve effector function, enhance cdc and/or adcc function, and improve stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
experimental examples
Example 1
Humanization of 2H7 Anti-CD20 Murine Monoclonal Antibody
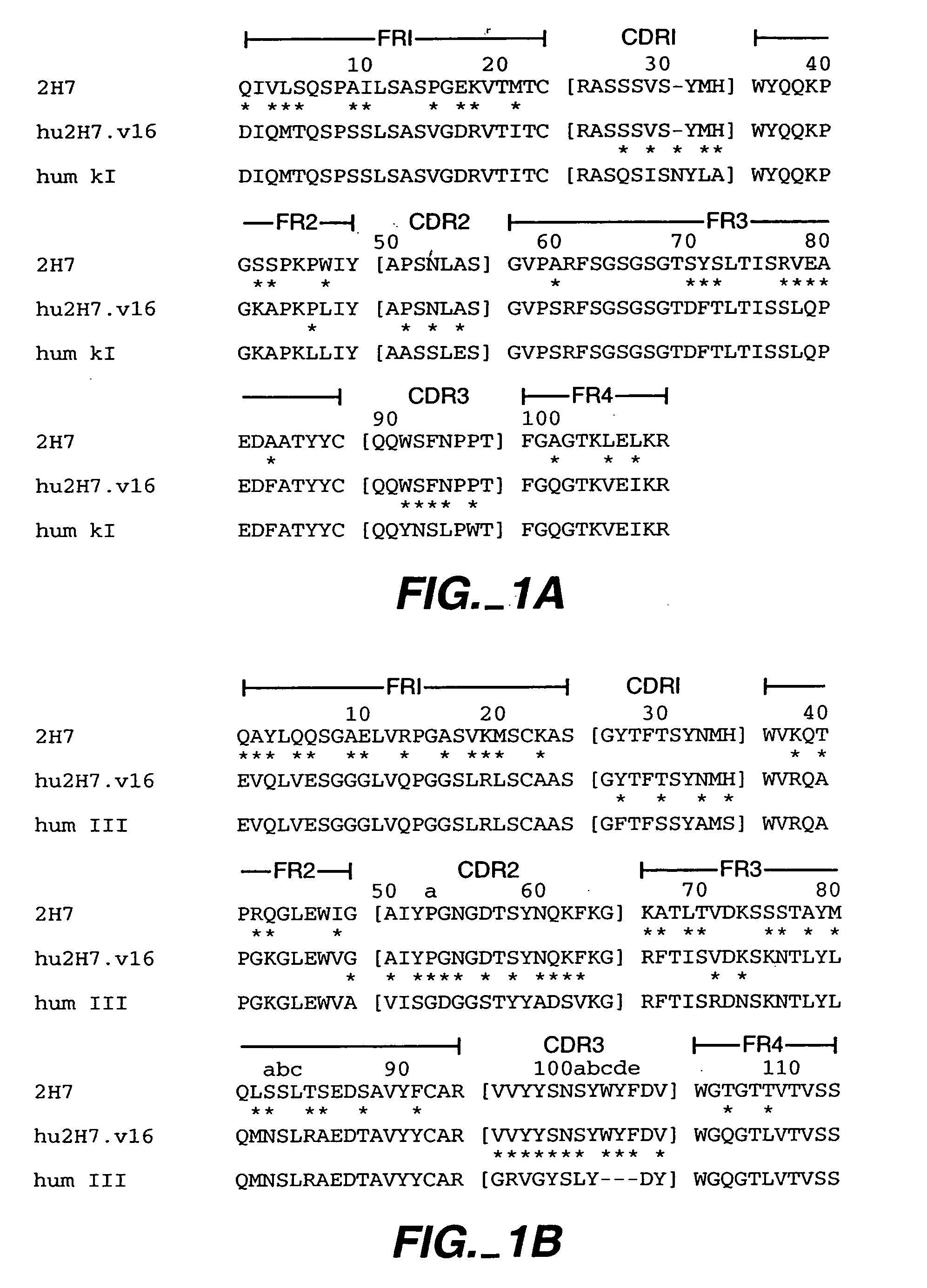
[0279] Humanization of the murine anti-human CD20 antibody, 2H7 (also referred to herein as m2H7, m for murine), was carried out in a series of site-directed mutagenesis steps. The murine 2H7 antibody variable region sequences and the chimeric 2H7 with the mouse V and human C have been described, see, e.g., U.S. Pat. Nos. 5,846,818 and 6,204,023. The CDR residues of 2H7 were identified by comparing the amino acid sequence of the murine 2H7 variable domains (disclosed in U.S. Pat. No. 5,846,818) with the sequences of known antibodies (Kabat et al., Sequences of proteins of immunological interest, Ed. 5. Public Health Service, National Institutes of Health, Bethesda, Md. (1991)). The CDR regions for the light and heavy chains were defined based on sequence hypervariability (Kabat et al., supra) and are shown in FIG. 1A and FIG. 1B, respectively. Using synthetic oligonucleotides (Table 1), site-directed mutagenesis (Kun...
example 2
Antigen-binding Determinants (paratope) of 2H7
[0292] Alanine substitutions (Cunningham & Wells, Science 244: 1081-1085 (1989) were made in 2H7.v16 or 2H7.v17 in order to test the contributions of individual side chains of the antibody in binding to CD20. IgG variants were expressed in 293 cells from pDR1 and pDR2 vectors, purified, and assayed for relative binding affinity as described above. Several alanine substitutions resulted in significant decreases in relative binding to CD20 on WIL-2S cells (Table 4).
TABLE 4Effects of alanine substitutions in the CDR regions of humanized 2H7.v16measured using cell-based ELISA (WIL2-S cells). The relative binding isexpressed as the concentration of the 2H7.v16 parent over theconcentration of the variant required for equivalent binding; hence aratio 1 indicateshigher affinity for the variant. Standard deviation in relative affinitydetermination averaged + / −10%. Framework substitutions in thevariable domains are relative to 2H7.v16 according...
example 3
Additional Mutations within 2H7 CDR Regions
[0293] Substitutions of additional residues and combinations of substitutions at CDR positions that were identified as important by Ala-scanning were also tested. Several combination variants, particularly v.96 appeared to bind more tightly than v.16.
TABLE 5Effects of combinations of mutations and non-alanine substitutionsin the CDR regions of humanized 2H7.v16 measured using cell-basedELISA (WIL2-S cells). The relative binding to CD20 is expressed asthe concentration of the 2H7.v16 parent over the concentration of thevariant required for equivalent binding; hence a ratio weaker affinity for the variant; a ratio >1 indicates higher affinityfor the variant. Standard deviation in relative affinity determinationaveraged + / −10%. Framework substitutions in the variabledomains are relative to 2H7.v16 according to the numbering systemof Kabat (Kabat et al., supra).2H7Heavy chainLight chainRelativeversionsubstitutionsSubstitutionsbinding16——-1-9...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com