Diagnostic assays for parvovirus B19
a parvovirus and diagnostic assay technology, applied in the field of parvovirus diagnostic assays, can solve the problems of unreliable serological diagnosis, impracticality of serological based tests, and difficult laboratory detection and isolation of the virus
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Parvovirus B19 Nucleic Acid Extraction for PCR
[0157] Human serum samples that had previously tested positive for human parvovirus B19 by either IgM or PCR tests were obtained from commercial sources and used to isolate DNA for subsequent PCR experiments. Samples were stored at −80° C. until used.
[0158] DNA was extracted from 0.2 mL of serum using the QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, Calif.) following the manufacturer's specifications with the following considerations. Carrier DNA was added to the lysis buffer to enhance nucleic acid binding and yield. In particular, an amount of 5.6 μg per sample of poly-adenylic acid 5′ (Sigma, St. Louis, Mo.) or poly-da (Roche, Indianapolis, Ind.) was added. Additionally, parvovirus B19 DNA was eluted with 200 μL of buffer AE (Qiagen) instead of water.
example 2
Detection of Parvovirus B19 Nucleic Acid-Positive Samples by PCR
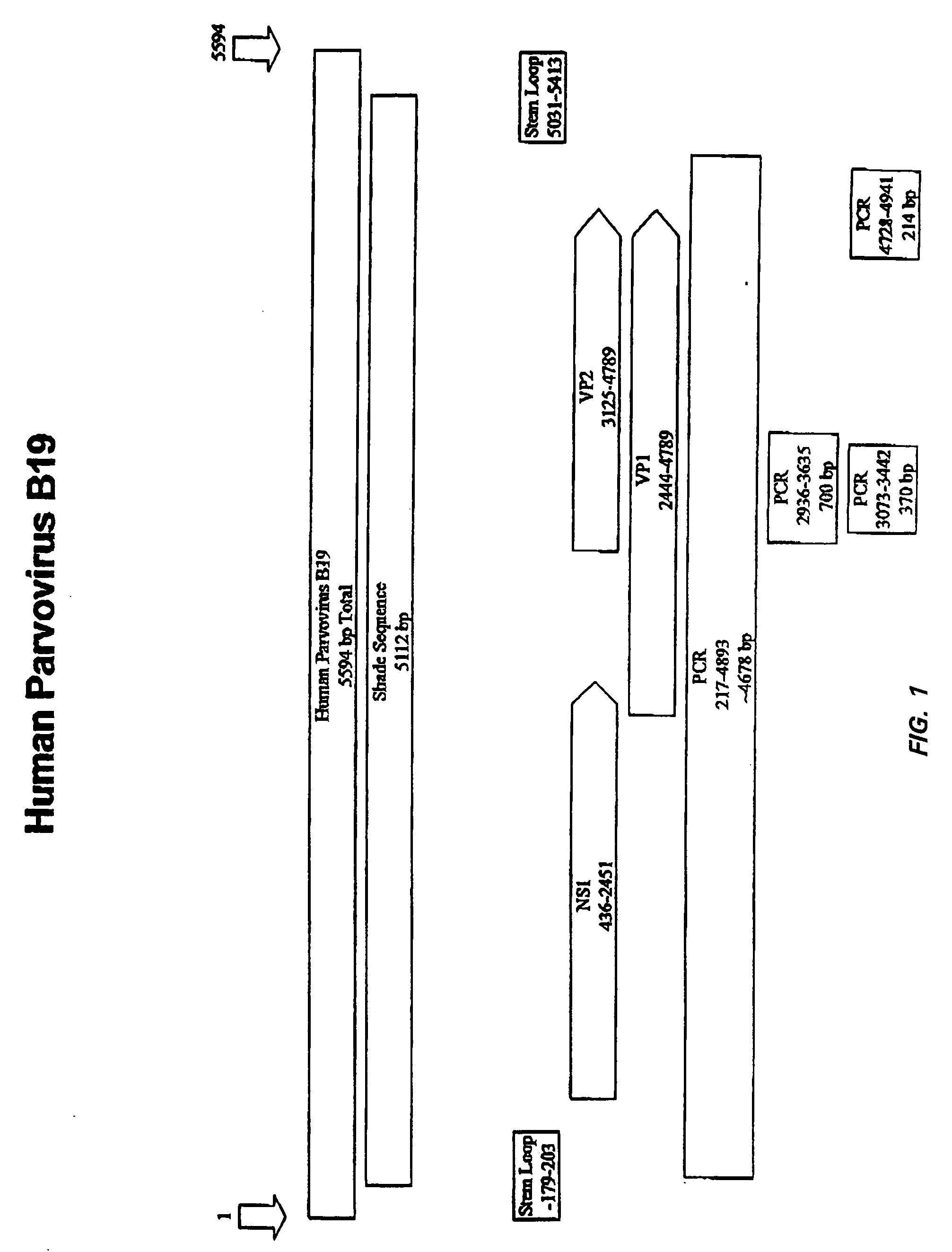
[0159] Two different PCR procedures were used to amplify parvovirus B19 fragments. One method, described in detail below, was used to amplify fragments of approximately 700 bp, 370 bp and 214 bp (see, FIG. 1). High Fidelity Expand PCR (Roche) was used to amplify fragments of approximately 4.7 kb. The approximately 700 bp fragment corresponds to nucleotide positions 2936-3635 of the parvovirus B19 genome described in Shade et al., J. Virol. (1986) 58:921-936. The approximately 370 bp occurs within the 700 bp fragment at nucleotide positions 3073-3442. The approximately 4.7 kb fragment is a 4677 nucleotide fragment corresponding to nucleotide positions 217-4893 of Shade et al., J. Virol. (1986) 58:921-936.
[0160] In order to amplify the B19 fragments of approximately 700 bp, 370 bp and 214 bp, the primers shown in Table 1 were used.
TABLE 1PCRGenomicPrimerSequenceproductregionVP-5AGGAAGTTTGCCGGAAGTTC370 bpVP1(SEQ ID NO:...
example 3
Cloning of Parvovirus B19 DNA Fragments
[0162] The PCR fragments were cloned into TOPO-TA vectors (Invitrogen, Carlsbad, Calif.). Cloning into these vectors is highly facilitated when the amplified DNA contains a single deoxyadenosine (A) at its 3′ end. Accordingly, a catalytic reaction to add the 3′ (A) overhead was used. The reaction mix contained 1.25 mM of dATP, 0.5 units of Taq polymerase (Perkin Elmer, Boston, Mass.) and proceeded at 72° C. for 15 min.
[0163] PCR fragments were cloned into the pCR2.1-TOPO vector using Invitrogen's TA cloning kit (TOPO™ TA CloningR Kit with One Shot TOP10 Electrocompetent Cells) following the manufacturer's specifications. Bacterial cells were incubated at 37° C. on Luria Broth plates containing ampicillin at 100 μg / ml, 0.66 mM IPTG and 0.033% X-Gal. A number of white colonies were inoculated in 4 mL of Luria-Broth ampicillin (100 μg / ml) and incubated overnight at 37° C. with shaking. Three mL of the overnight cultures were used to prepare plas...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com