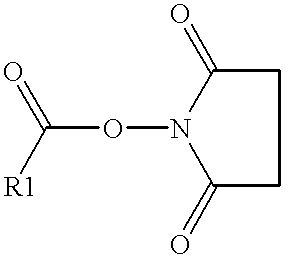
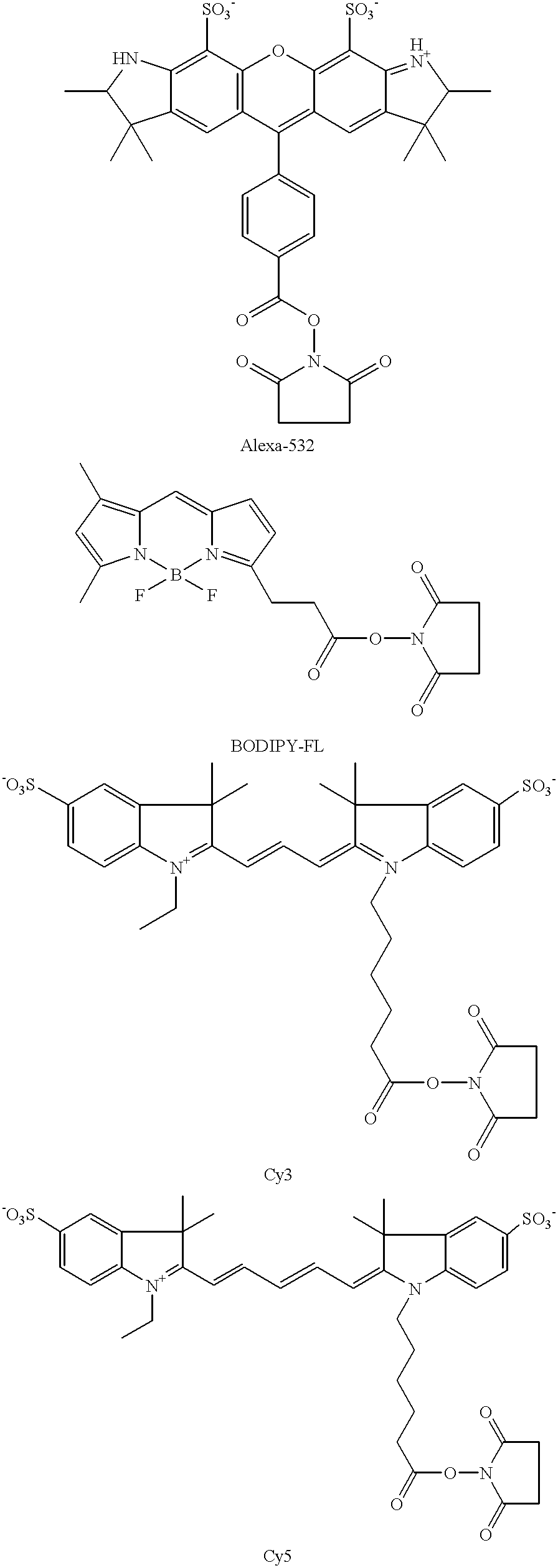
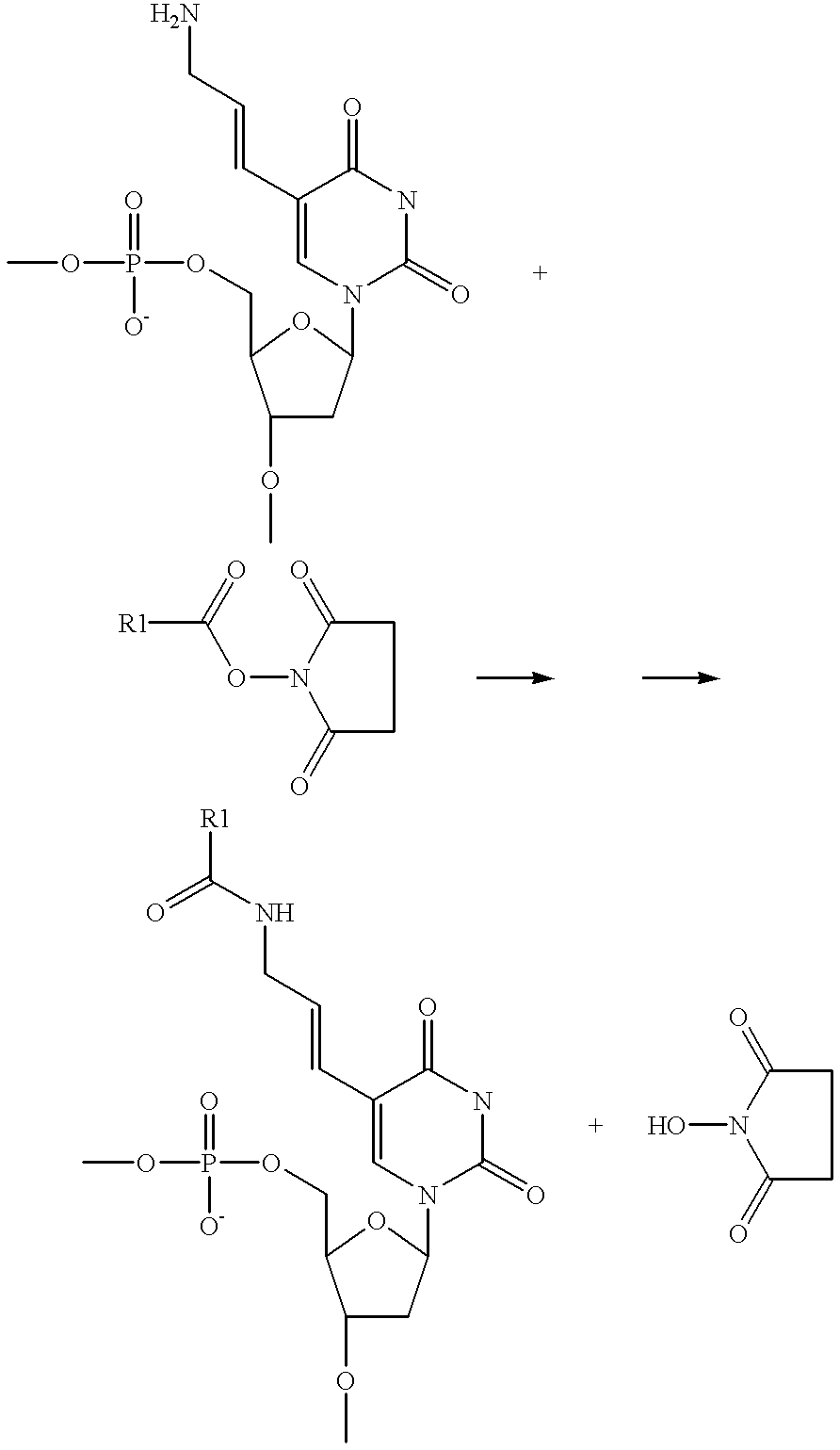
Methods of labeling nucleic acids for use in array based hybridization assays
a nucleic acid and array technology, applied in the field of nucleic acid labeling, can solve the problems of limited spectrum of fluorescent labels that may be employed in protocols where labeled nucleotide analogs are generated from labeled nucleotide analogs, and not per
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0051] Preparation of Cy3 labeled target nucleic acid.
[0052] 1. To 5 .mu.g of polyA.sup.+ placental RNA add 5 .mu.l of gene specific primers (0.2 .mu.M each as described in PCT / US98 / 10561 the disclosure of which is herein incorporated by reference). Add water to total final volume of 25 .mu.l.
[0053] 2. Heat at 70.degree. C. for 5 min.
[0054] 3. Cool to 48.degree. C., add 25 .mu.l of following master mix:
1 5 .times. first strand buffer 10 .mu.l 10 .times. dNTP mixture 5 .mu.l MMLV RT (200 u / ml) 2.5 .mu.l Milli Q water 7.5 .mu.l
[0055]
2 Composition of 10 .times. dNTP mixture for 100 .mu.l Reagent Stock conc. Amount Final conc. DATP 100 mM 5 .mu.l 5 mM DCTP 100 mM 5 .mu.l 5 mM DGTP 100 mM 5 .mu.l 5 mM DTTP 100 mM 2.5 .mu.l 2.5 mM Allylamino-dUTP 10 mM 25 .mu.l 2.5 mM Milli Q water 57.5 .mu.l
[0056] 4. Incubate at 48.degree. C. for 30 min.
[0057] 5. Heat to 70.degree. C. for 5 min.
[0058] 6. Cool to 37.degree. C. and add 0.5 .mu.l of RNase H (10 U / .mu.l)
[0059] 7. Incubate at 37.degree. C. fo...
example 2
[0079] Post Hybridization Labeling
[0080] Target nucleic acids are prepared as described above, except that the amino functional group is replaced with an SH group. The target nucleic acids are then hybridized to an array of probe nucleic acids. Following hybridization, the array surface is washed to removed unhybridized target. The array surface is then contacted with a solution of maleimide functionalized label under conditions sufficient for the functionalized label to conjugate to the hybridized target via the SH moiety, resulting in an array of labeled hybridized targets.
[0081] It is evident that the subject invention provides an important new method for generating labeled target nucleic acids for use in array based hybridization assays. With the subject methods, one is not limited to using labeled analogs that can be processed by polymerases, since the labeling step occurs after the target generation step. As such, the number of different types of labels that can now be used in...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com