Quick-speed PCR quantitative determination method of sulfate-reducing bacteria direct dilution by multiple proportions
A quantitative detection method and multiple dilution technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of long detection cycle, inability to guide production in real time, and high detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
[0005] Specific embodiment 1: This embodiment is a method for rapid quantitative detection of sulfate-reducing bacteria by direct doubling dilution PCR, which sequentially includes the following steps: (1). Preparation of cell pretreatment buffer; (2). Bacterial cell Preparation; (3). Time ratio dilution; (4). PCR operation on ice; (5). PCR amplification on a thermal cycler; (6). 1.0% agarose electrophoresis detection; (7) Observe the results, check the table and count; the PCR reaction system in the process of "(4). Perform PCR operation on ice" is 20μl: including: Buffer (10×) 2μl, 0.3mmol·L -1 D NTP 2μl, concentration is 0.1μmol·L -1 The forward primer SRBF 1μl, the concentration is 0.1μmol·L -1 1μl of reverse primer SRBR, 0.3U rTaq enzyme, 11.7μl of deionized water for the rest, and then 2μl of sample.
Example Embodiment
[0006] Specific implementation manner 2: The detailed operation process of this implementation manner is as follows:
[0007] (1). Preparation of cell pretreatment buffer: the mother liquor components of "Bacterial cell pretreatment buffer" are: 70g NaCi, 2g Kci, 12g Na 2 HPO 4 , 2g KH 2 PO 4 , 1‰ SDS; Dilute the mother liquor 10 times to become the pretreatment buffer of the bacteria; control the pH of the working solution to 7.5 and sterilize it at 121°C for 15-30 minutes.
[0008] (2). Preparation of bacterial cells: The following steps are included in sequence: a. The preparation of bacterial cells is completed under an ultra-clean workbench, which needs to be sterilized by ultraviolet light for 20-40 minutes, and then used with high-purity nitrogen Take 1mL of the oilfield sewage water sample retrieved by the anaerobic tube or bioreactor sludge and inject 1mL into the sterilized 1.5mL centrifuge tube; b. Fully shake the oilfield sewage water sample on the shaker 3~6min, fully...
Example Embodiment
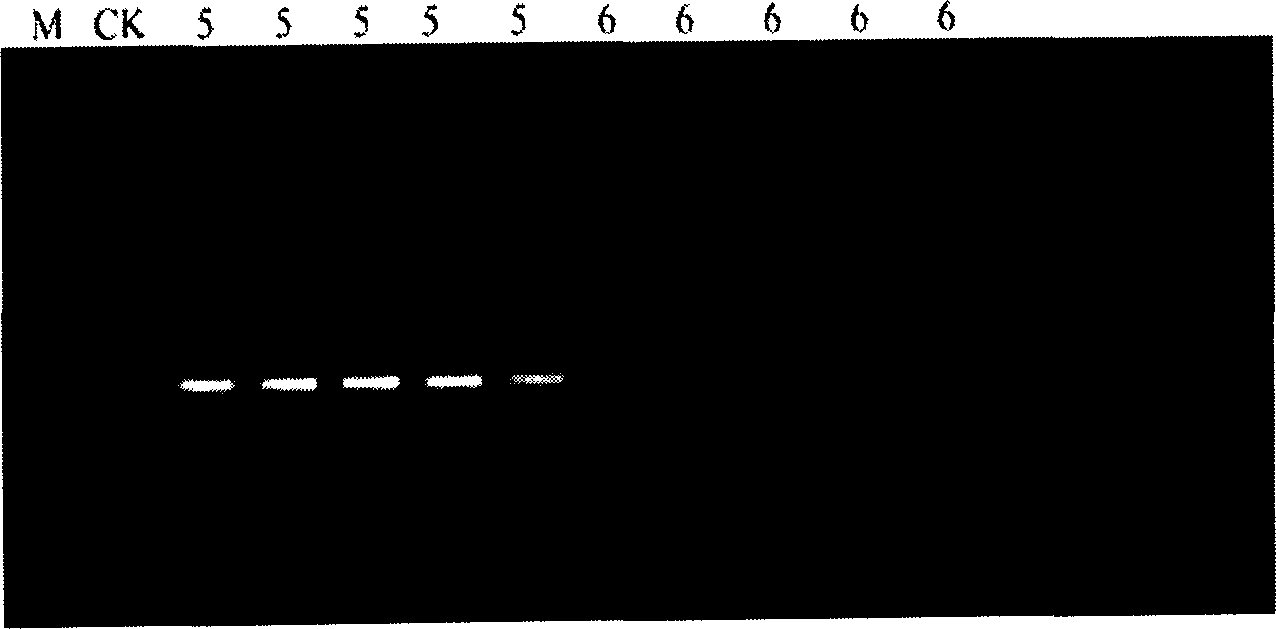
[0014] Specific embodiment 3: In this embodiment, the oilfield sewage retrieved by the anaerobic tube containing high-purity nitrogen is used as the test sample, and the five-tube parallel method is used to perform the PCR operation on ice, with 5 samples for each gradient. 1.0% agarose electrophoresis detection, the detection result is as follows figure 1 , figure 2 Shown. Observe the results, check the meter and count.
[0015] The specific look-up table calculation method is: select the last three serial dilution gradients (10 x , 10 x+1 , 10 x+2 ), the number of tubes with bacterial DNA bands is arranged in the order of the dilution gradient from small to large into a three-digit number called the band approximate value (abc). Among them, the first dilution gradient (10 x ) Is the last dilution gradient with bacterial DNA bands in the last 5 test tubes. If the third dilution gradient (10 x+2 ) There are still bacterial DNA bands appearing in the next gradient dilution tube, t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com