Method and device for structural variation detection and storage medium
A technology of structural variation and signal, applied in the field of bioinformatics, can solve the problems of accuracy impact, difficult interchromosomal structure, time-consuming, etc., achieve high detection rate and precision, and reduce the effect of false positive signals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
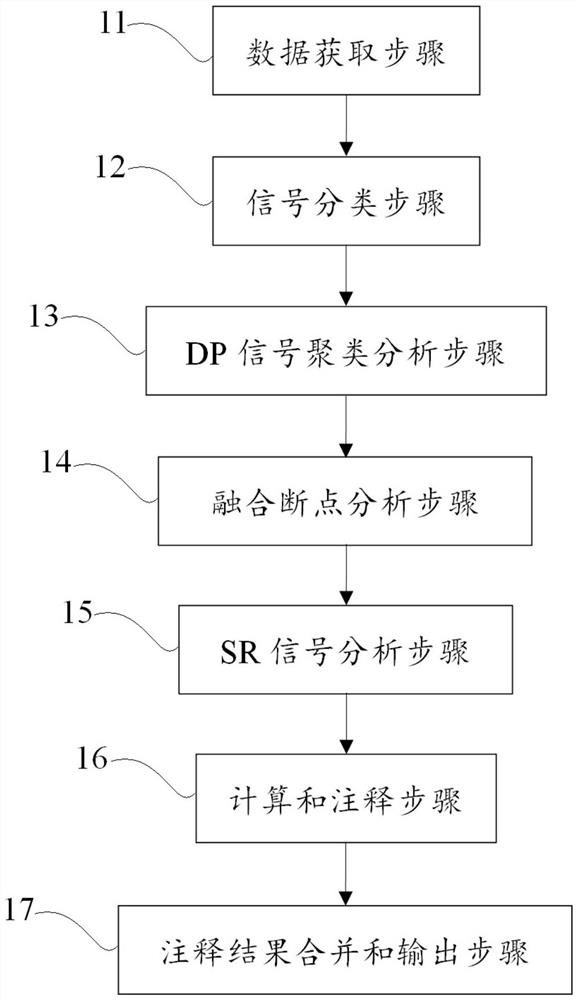
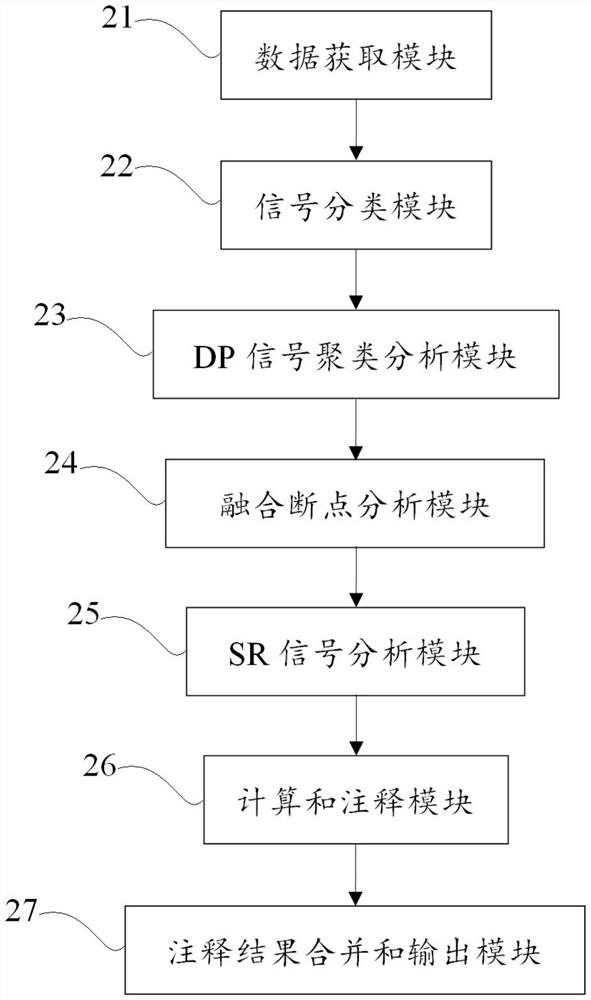
[0087] The structural variation detection method in this example is as follows:
[0088] Input: preprocessed bam file, reference series;
[0089] 1. Data acquisition steps, including acquiring the bam file of the next-generation sequencing data of the object to be tested, calculating the basic information of the bam file, insert size mean and standard deviation, insert size max (insert size mean+3.96*insertsize std), and reads length;
[0090] 2. In the signal classification step, the reads in the interval are extracted in parallel according to the length of 75k from the bam file, and the abnormal reads are divided into four signals: DP (insert size>insert size max or two paired reads fall on two different chromosomes ), SR (reads with soft shearing), SU (only one of the read pairs matches the reference sequence), and put them in a temporary file after extraction;
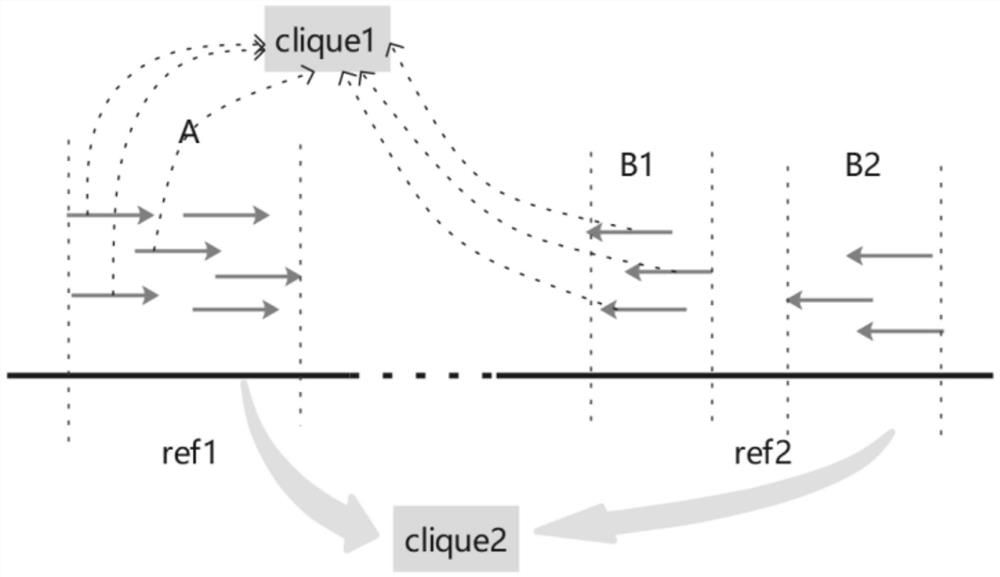
[0091] 3. DP signal clustering analysis step, cluster the DP signals extracted in step 2, find DP signal cluste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com