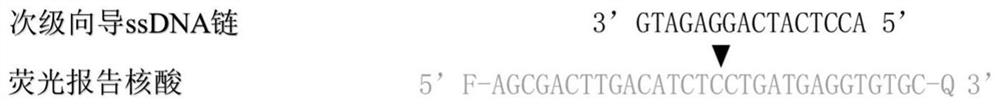Nucleic acid detection method based on normal-temperature prokaryotic Argonaute protein and application thereof
A normal temperature, protein technology, applied in the biological field, can solve the problems of complex design principles, high cost, and long time consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0191] Preparation and detection method of nucleic acid detection reagent based on PbAgo
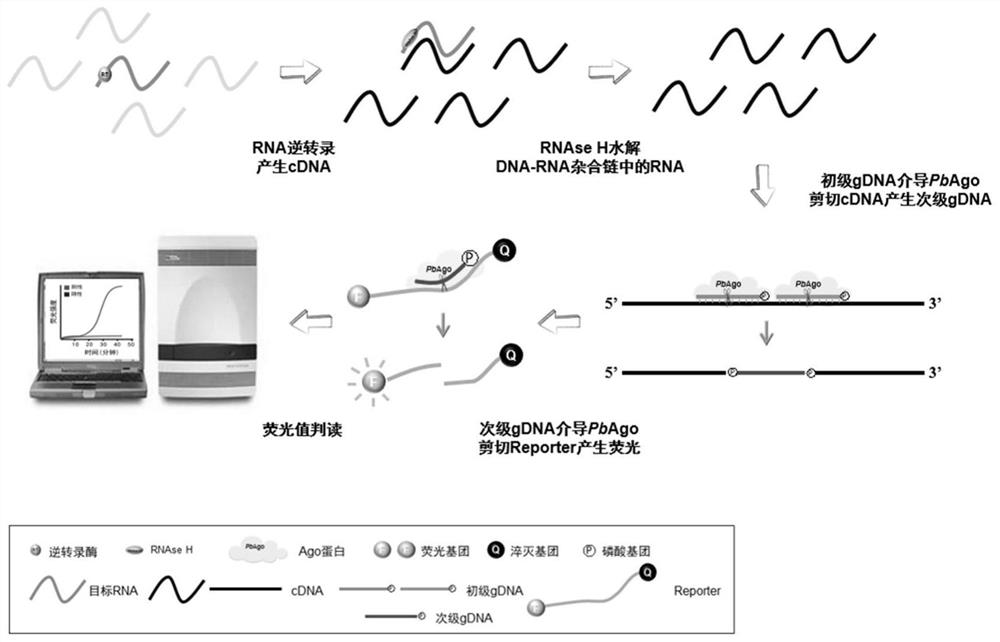
[0192] In this embodiment, the reagents used in the nucleic acid detection method based on the gene editing enzyme Paenibacillus borealisArgonaute (PbAgo) of the present invention and their usage methods are provided.
[0193] 1.1 Detection reagent
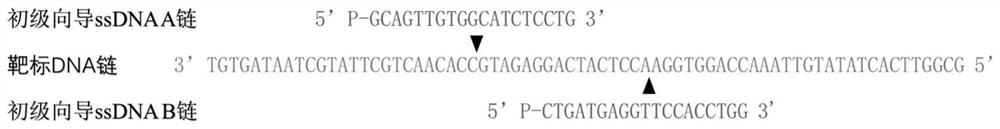
[0194] 在本实施例中,以检测SRAS-COV2 1b基因为例子,相应的特异性目标核酸序列为5’-GGGGAUAAAAGUGCAUUAACAUUGGCCGUGACAGCUUGACAAAUGU UAAAAACACUAUUAGCAUAAGCAGUUGUGGCAUCUCCUGAUGAGGUUC CACCUGGUUUAACAUAUAGUGAACCGCCACACAUGACCAUUUCACUCA AUACUUGAGCACACUCAUUAGCUAAUCUAUAGAAACGGUGUGACAAGC UACAACACGUUGU-3’,SEQ ID No.:1。
[0195] Based on the method of the present invention, the corresponding detection reagents include the following:
[0196] (1), reverse transcription primer R-primer, the specific sequence is as follows:
[0197] R-primer: 5'-ACAACGTGTTGTAGC-3' (SEQ ID No.2)
[0198] (2), specific guide ssDNAs pair, including sense strand guide ssDNA and nonsense strand guide ssDNA, ...
Embodiment 2
[0228] Preparation and detection method of nucleic acid detection reagent based on KmAgo
[0229] In this embodiment, the reagents used in the nucleic acid detection method based on the gene editing enzyme Kurthia massiliensis Argonaute (KmAgo) of the present invention and their usage methods are provided.
[0230] 2.1 Detection reagent
[0231] 在本实施例中,以检测SRAS-COV2 1b基因为例子,相应的特异性目标核酸序列为5’-GGGGAUAAAAGUGCAUUAACAUUGGCCGUGACAGCUUGACAAAUGU UAAAAACACUAUUAGCAUAAGCAGUUGUGGCAUCUCCUGAUGAGGUUC CACCUGGUUUAACAUAUAGUGAACCGCCACACAUGACCAUUUCACUCA AUACUUGAGCACACUCAUUAGCUAAUCUAUAGAAACGGUGUGACAAGC UACAACACGUUGU-3’,SEQ ID No.:1。
[0232] Based on the method of the present invention, the corresponding detection reagents include the following:
[0233] (1), reverse transcription primer R-primer, the specific sequence is as follows:
[0234] R-primer: 5'-ACAACGTGTTGTAGC-3' (SEQ ID No.2)
[0235] (2), specific guide ssDNAs pair, including sense strand guide ssDNA and nonsense strand guide ssDNA, t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com