Biomarker for esophageal cancer typing and application thereof
A biomarker, esophageal cancer technology, applied in the field of medical detection, can solve the problems of poor differentiation, wrong pathological determination, low qualification rate, etc., and achieve the effect of ensuring accurate, convenient and precise treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Based on the TCGA public database, a preliminary screening of variant gene markers for pathological subtypes of esophageal cancer was performed.
[0044] The screening method is as follows.
[0045] 1. Obtain the whole exome sequencing data of the tumor tissue of patients with esophageal cancer from the TCGA database.
[0046] In this example, the whole exome sequencing data of 184 patients with esophageal cancer (including 88 cases of adenocarcinoma and 96 cases of squamous cell carcinoma) were downloaded, and seven different software were used: MuTect, VarScan, MuSE, Radia and SomaticSniper to detect SNPs respectively Mutation site: VarScan, Pindel, and Indelocator were used to detect InDels respectively, and finally the mutation site that was simultaneously called by at least two mutation software in the sample was used as the candidate mutation site.
[0047] 2. According to the data set of adenocarcinoma group and the data set of squamous cell carcinoma group, the...
Embodiment 2
[0057] For the potential markers obtained in Example 1, clinical samples were analyzed and verified.
[0058] The verification process is as follows.
[0059] 1. Acquisition of tissue samples: 184 cases of esophageal cancer (96 cases of squamous cell carcinoma, 88 cases of squamous cell carcinoma) related FFPE section samples were collected from Jinan University.
[0060] 2. Sample sequencing analysis: FFPE tissue samples were subjected to whole-genome sequencing analysis by a third party (Brightcode Biotechnology Company).
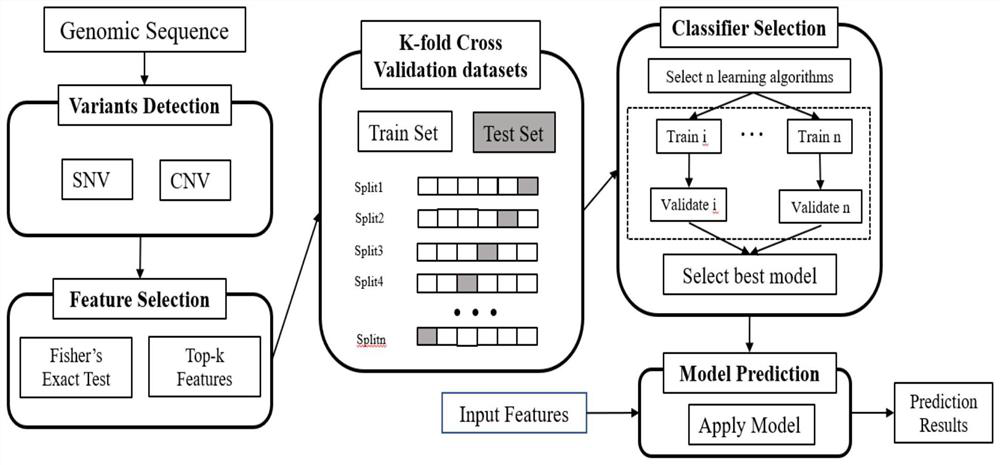
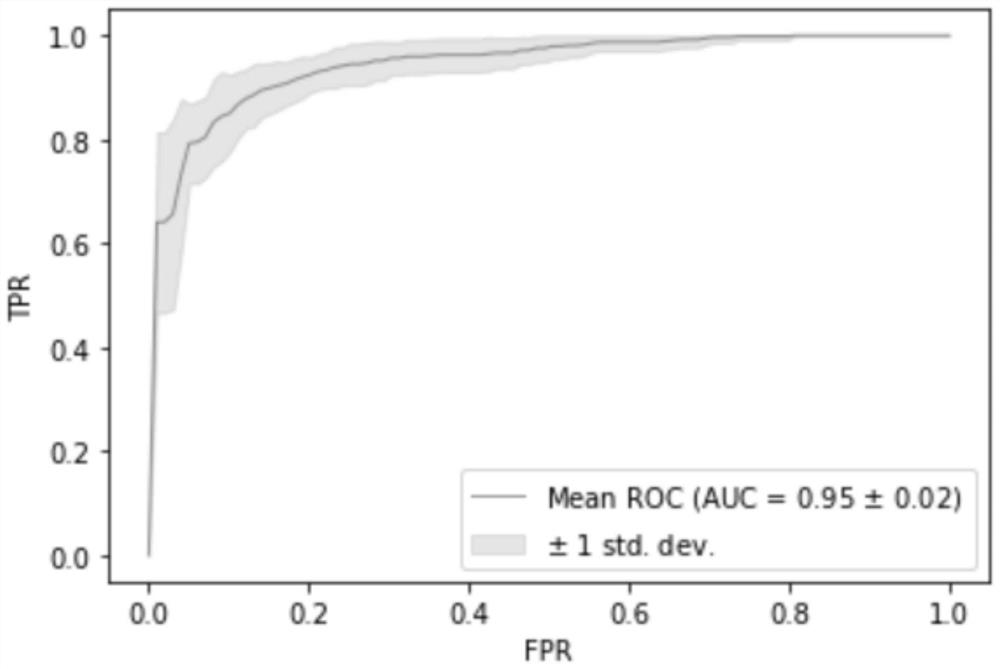
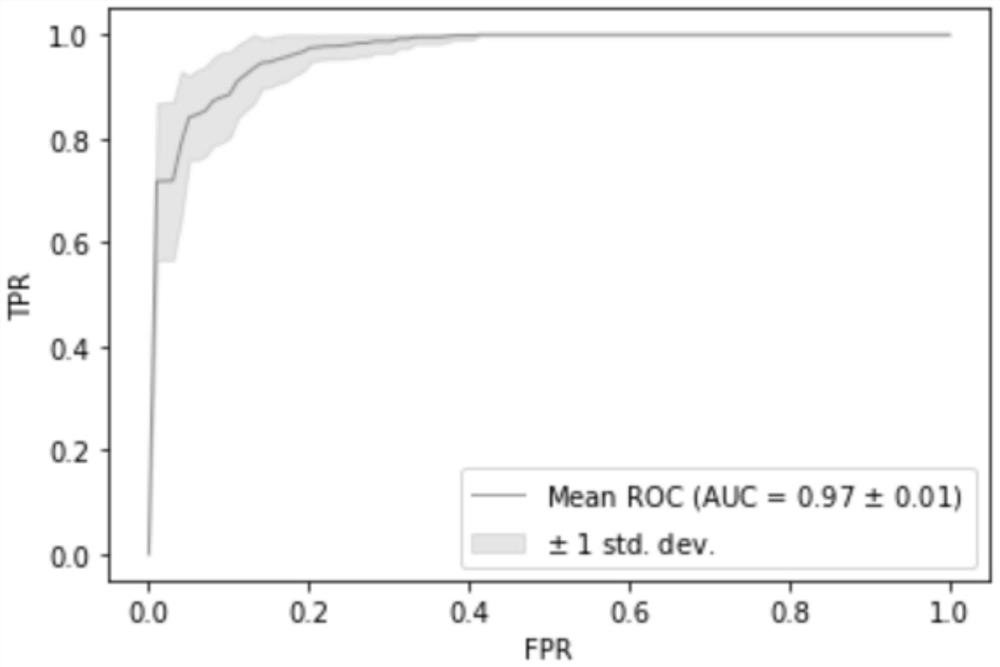
[0061] 3. Establishing a model: using the information of all target markers in Example 1, the independent verification set, namely 96 cases of squamous cell carcinoma and 88 cases of adenocarcinoma patient tissue samples were detected and judged, and the random forest model was used for modeling analysis. Modeling process such as figure 1 As shown, according to the 7:3 segmentation, 20 repetitions, the AUC of the model is as high as 0.9489, and the mode...
Embodiment 3
[0066] Select 28 samples clinically judged as esophageal cancer, use the 10 MARKER combination models established in the above-mentioned Example 2 for analysis, and compare the analysis results with the judgment results of clinical experts, the results are shown in the table below.
[0067] Table 3 clinical verification results
[0068]
[0069]
[0070] It can be seen from the above results that using the biomarkers of the present invention, the above model can be used to determine the classification of squamous cell carcinoma or adenocarcinoma in lung small cell carcinoma more accurately, and the consistency with expert judgment is more than 91.3%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com