Primer set and method for identifying Streptomyces flavus w68
A technology of Streptomyces albus and primer set, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/inspection, achieving the effects of high detection efficiency, high sensitivity, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] material method
[0034] Streptomyces albidoflavus W68 is isolated from marine sludge. It is a Streptomyces strain with significant biocontrol effect on plant fungal diseases. It can be used in microbial fertilizers or microbial pesticides. It has been preserved on March 14, 2016. General Microbiology Center of China Microbial Culture Collection Management Committee, the preservation number is CGMCC No. 12210, the address of the preservation unit is No. 3, No. 1, Beichen West Road, Chaoyang District, Beijing, and the preservation unit is abbreviated as CGMCC.
[0035] The genome sequence of Streptomyces flavus W68 has been submitted to the NCBI database on November 19, 2020, and the GenBank number is: CP064783.1. The genome size of this strain is 6796629 bp.
[0036] PCR amplification system: 10× buffer (10×Taq Buffer) 2uL, 2.5mM deoxynucleotide triphosphates (dNTPs) 1.6uL, 10uM forward primer and reverse primer each 0.8uL, DNA polymerase (TaqDNA polymerase) 0.2uL, te...
Embodiment 2
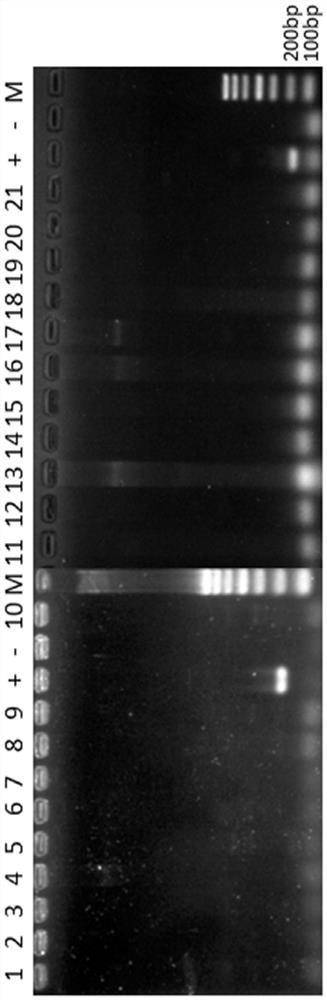
[0045] Screening of DNA Molecular Markers in Genome Sequence of Streptomyces flavinus W68
[0046] Based on the complete genome sequence of Streptomyces lividans W68, the following steps were taken to screen specific sequences:
[0047] Cut the genome sequence into 300bp window bins;
[0048] Compare the obtained bins sequence with the bacterial genome database;
[0049] Filter out bins whose alignment length is greater than 100 bp and whose homology is greater than 80%;
[0050] Merge the filtered consecutive bins.
[0051] Through the above steps, 232 nucleic acid sequence fragments of different lengths were obtained;
[0052] Further, the above 232 nucleic acid sequences were manually screened to obtain 10 target sequences for verification;
[0053] Primers were designed for the above 10 nucleic acid sequences using primer6 software, PCR amplification was performed on different templates, and the following two specific nucleic acid fragments were finally screened.
Embodiment 3
[0055] Design and detection of primers specific for sequences 1751401-1751700
[0056] Using the primer6 software, after inputting the nucleic acid sequence fragments 1751401-1751700, the primer sets P1751437F and P1751612R were obtained. The relevant information of the primers is shown in Table 2.
[0057] Table 2: Indexes of primer sets P1751437F and P1751612R
[0058]
[0059] After the primer sets P1751437F and P1751612R were added to the PCR amplification system in Example 1 according to the proportions, an ordinary PCR amplification reaction was carried out according to the PCR amplification procedure in Example 1.
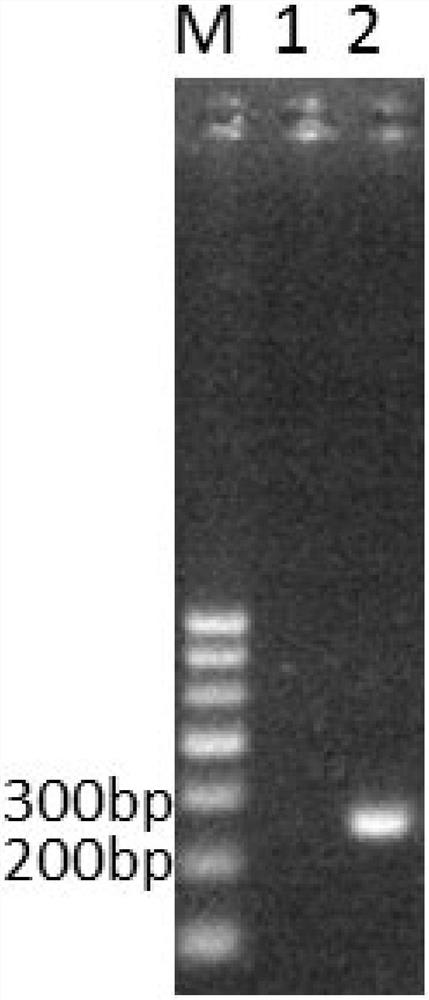
[0060] like figure 1 As shown, M stands for Marker, and its fragment sizes from top to bottom are 700bp, 600bp, 500bp, 400bp, 300bp, 200bp, 100bp; the lane number 1 is ddH 2 O is the template, and no amplification product was detected; the swimming lane of No. 2 is the genome of Streptomyces lividans W68 as the template, after the PCR amplification reac...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap